2CZY
 
 | | Solution structure of the NRSF/REST-mSin3B PAH1 complex | | Descriptor: | Paired amphipathic helix protein Sin3b, transcription factor REST (version 3) | | Authors: | Nomura, M, Uda-Tochio, H, Murai, K, Mori, N, Nishimura, Y. | | Deposit date: | 2005-07-20 | | Release date: | 2005-12-20 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The Neural Repressor NRSF/REST Binds the PAH1 Domain of the Sin3 Corepressor by Using its Distinct Short Hydrophobic Helix
J.Mol.Biol., 354, 2005
|
|
2JZ4
 
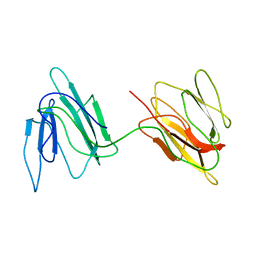 | | Putative 32 kDa myrosinase binding protein At3g16450.1 from Arabidopsis thaliana | | Descriptor: | Jasmonate inducible protein isolog | | Authors: | Takeda, N, Sugimori, N, Torizawa, T, Terauchi, T, Ono, A.M, Yagi, H, Yamaguchi, Y, Kato, K, Ikeya, T, Guntert, P, Aceti, D.J, Markley, J.L, Kainosho, M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-02-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the putative 32 kDa myrosinase-binding protein from Arabidopsis (At3g16450.1) determined by SAIL-NMR.
Febs J., 275, 2008
|
|
1NHU
 
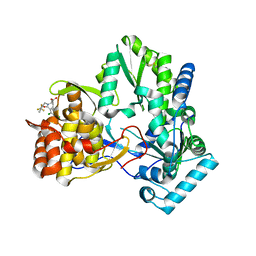 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(2,4-DICHLORO-BENZOYL)-(3-TRIFLUOROMETHYL-BENZYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
1NHV
 
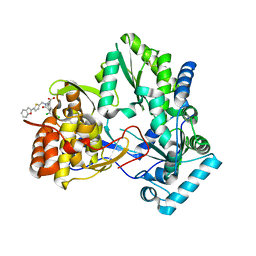 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(5-BENZOFURAN-2-YL-THIOPHEN-2-YLMETHYL)-(2,4-DICHLORO-BENZOYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
4Y9H
 
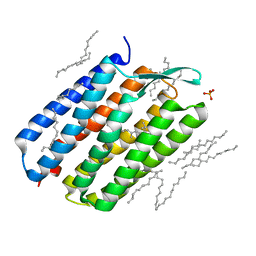 | | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles | | Descriptor: | Bacteriorhodopsin, DECANE, DODECANE, ... | | Authors: | Saiki, H, Sugiyama, S, Kakinouchi, K, Kawatake, S, Hanashima, S, Matsumori, N, Murata, M. | | Deposit date: | 2015-02-17 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles
To Be Published
|
|
7BYD
 
 | | Crystal structure of SN45 TCR in complex with lipopeptide-bound Mamu-B*05104 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, B protein, ... | | Authors: | Morita, D, Sugita, M, Iwashita, C. | | Deposit date: | 2020-04-22 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.80003262 Å) | | Cite: | Crystal structure of the ternary complex of TCR, MHC class I and lipopeptides.
Int.Immunol., 32, 2020
|
|
6IWH
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with C14-GGGI lipopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, C14-GGGI lipopeptide, ... | | Authors: | Yamamoto, Y, Morita, D, Sugita, M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification and Structure of an MHC Class I-Encoded Protein with the Potential to PresentN-Myristoylated 4-mer Peptides to T Cells.
J Immunol., 202, 2019
|
|
6IWG
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with N-myristoylated 4-mer lipopeptide derived from SIV nef protein | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, Beta-2-microglobulin, ... | | Authors: | Yamamoto, Y, Morita, D, Sugita, M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Structure of an MHC Class I-Encoded Protein with the Potential to PresentN-Myristoylated 4-mer Peptides to T Cells.
J Immunol., 202, 2019
|
|
7WJ3
 
 | | Crystal structure of HLA-C*1402 complexed with 4-mer lipopeptide | | Descriptor: | 1,2-ETHANEDIOL, 4-mer lipopeptide, Beta-2-microglobulin, ... | | Authors: | Kuroha, J, Morita, D, Asa, M, Sugita, M. | | Deposit date: | 2022-01-05 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56343615 Å) | | Cite: | Crystal structures of N-myristoylated lipopeptide-bound HLA class I complexes indicate reorganization of B-pocket architecture upon ligand binding.
J.Biol.Chem., 298, 2022
|
|
7WT4
 
 | | Crystal structure of HLA-A*2402 complexed with 8-mer Influenza PB1 peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Asa, M, Morita, D, Sugita, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89459145 Å) | | Cite: | Crystal structures of N-myristoylated lipopeptide-bound HLA class I complexes indicate reorganization of B-pocket architecture upon ligand binding.
J.Biol.Chem., 298, 2022
|
|
7WT5
 
 | | Crystal structure of HLA-A*2450 complexed with 8-mer model peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 8-mer model peptide, ... | | Authors: | Asa, M, Morita, D, Sugita, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.0950768 Å) | | Cite: | Crystal structures of N-myristoylated lipopeptide-bound HLA class I complexes indicate reorganization of B-pocket architecture upon ligand binding.
J.Biol.Chem., 298, 2022
|
|
7WJ2
 
 | | Crystal structure of HLA-C*1402 complexed with 8-mer HIV gag peptide | | Descriptor: | 1,2-ETHANEDIOL, 8-mer peptide, ACETATE ION, ... | | Authors: | Kuroha, J, Morita, D, Asa, M, Sugita, M. | | Deposit date: | 2022-01-05 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structures of N-myristoylated lipopeptide-bound HLA class I complexes indicate reorganization of B-pocket architecture upon ligand binding.
J.Biol.Chem., 298, 2022
|
|
7WT3
 
 | | Crystal structure of HLA-A*2402 complexed with 4-mer lipopeptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-mer lipopeptide, ... | | Authors: | Asa, M, Morita, D, Sugita, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.887655 Å) | | Cite: | Crystal structures of N-myristoylated lipopeptide-bound HLA class I complexes indicate reorganization of B-pocket architecture upon ligand binding.
J.Biol.Chem., 298, 2022
|
|
6LB2
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with mono-acyl glycerol | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2019-11-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69380951 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
6LAH
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with lysophosphatidylcholine | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
6LAM
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with lysophosphatidylethanolamine | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
6LT6
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with lysophosphatidylcholine | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
5B34
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles in Complex with Iodine-labeled Detergent HAD13a Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | 2,4,6-tris(iodanyl)-5-(octanoylamino)benzene-1,3-dicarboxylic acid, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5B35
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T, Suzuki, M. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4ZFZ
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with myristoylated 5-mer lipopeptide derived from SIV Nef protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-mer lipopeptide from Protein Nef, ... | | Authors: | Morita, D, Sugita, M. | | Deposit date: | 2015-04-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal structure of the N-myristoylated lipopeptide-bound MHC class I complex
Nat Commun, 7, 2016
|
|
6ITD
 
 | | Crystal structure of BioU (K124A) from Synechocystis sp.PCC6803 in complex with the analog of reaction intermediate, 3-(1-aminoethyl)-nonanedioic acid | | Descriptor: | 3-(1-AMINOETHYL)NONANEDIOIC ACID, Slr0355 protein | | Authors: | Sakaki, K, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2018-11-21 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
6IR4
 
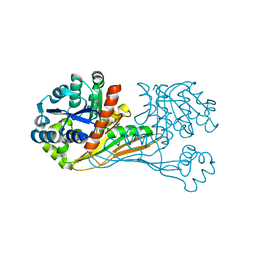 | | Crystal structure of BioU from Synechocystis sp.PCC6803 (apo form) | | Descriptor: | Slr0355 protein | | Authors: | Sakaki, K, Oishi, K, Shimizu, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2018-11-10 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
6K36
 
 | |
5GUE
 
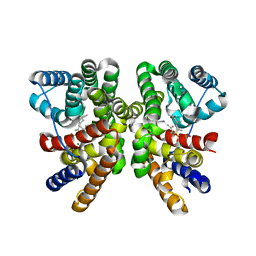 | | Crystal structure of CotB2 (GGSPP/Mg2+-Bound Form) from Streptomyces melanosporofaciens | | Descriptor: | Cyclooctat-9-en-7-ol synthase, MAGNESIUM ION, phosphonooxy-[(10E)-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraenyl]sulfanyl-phosphinic acid | | Authors: | Tomita, T, Kim, S.-Y, Ozaki, T, Yoshida, A, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2016-08-28 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the CotB2-Catalyzed Cyclization of Geranylgeranyl Diphosphate to the Diterpene Cyclooctat-9-en-7-ol
ACS Chem. Biol., 12, 2017
|
|
5GUC
 
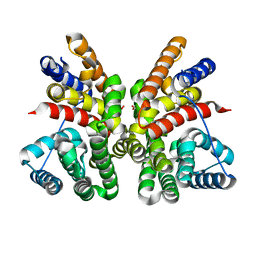 | | Crystal structure of CotB2 (apo form) from Streptomyces melanosporofaciens | | Descriptor: | Cyclooctat-9-en-7-ol synthase, FORMIC ACID | | Authors: | Tomita, T, Kim, S.-Y, Ozaki, T, Yoshida, A, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2016-08-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the CotB2-Catalyzed Cyclization of Geranylgeranyl Diphosphate to the Diterpene Cyclooctat-9-en-7-ol
ACS Chem. Biol., 12, 2017
|
|
