5DA6
 
 | |
4RBQ
 
 | | 32 base pair oligo(U) RNA | | Descriptor: | POTASSIUM ION, U-Helix RNA from Trypanosome editing | | Authors: | Mooers, B.H.M. | | Deposit date: | 2014-09-12 | | Release date: | 2015-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure of the Trypanosome RNA Editing U-Helix with 16 Contiguous Us
To be Published
|
|
3FI5
 
 | | Crystal Structure of T4 Lysozyme Mutant R96W | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Lysozyme, ... | | Authors: | Mooers, B.H.M, Matthews, B.W. | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3F9L
 
 | |
3F8V
 
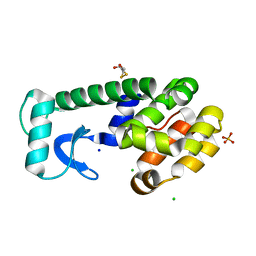 | |
3FA0
 
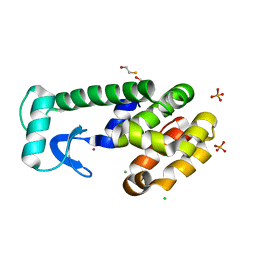 | |
3FAD
 
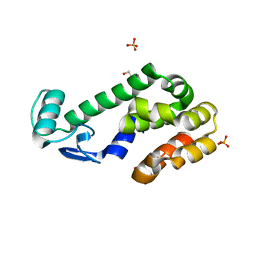 | |
3ND3
 
 | | Uhelix 16-mer dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*GP*AP*AP*GP*AP*UP*UP*UP*UP*UP*UP*UP*U)-3', POTASSIUM ION, SODIUM ION | | Authors: | Mooers, B.H, Singh, A. | | Deposit date: | 2010-06-07 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The crystal structure of an oligo(U):pre-mRNA duplex from a trypanosome RNA editing substrate.
Rna, 17, 2011
|
|
3ND4
 
 | | Watson-Crick 16-mer dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*GP*AP*AP*GP*AP*UP*CP*UP*UP*CP*UP*CP*U)-3', MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Mooers, B.H, Singh, A. | | Deposit date: | 2010-06-07 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.524 Å) | | Cite: | The crystal structure of an oligo(U):pre-mRNA duplex from a trypanosome RNA editing substrate.
Rna, 17, 2011
|
|
1SWY
 
 | |
1SX2
 
 | |
1SX7
 
 | |
1SWZ
 
 | |
5D99
 
 | | 3DW4 redetermined by direct methods starting from random phase angles | | Descriptor: | GLYCEROL, RNA (27-MER) hairpin from sarcin-ricin domain of E. coli 23S rRNA | | Authors: | Mooers, B.H.M. | | Deposit date: | 2015-08-18 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Direct-methods structure determination of a trypanosome RNA-editing substrate fragment with translational pseudosymmetry.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3C83
 
 | |
3C81
 
 | |
3C8S
 
 | |
3C7W
 
 | |
3C7Z
 
 | | T4 lysozyme mutant D89A/R96H at room temperature | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3C8Q
 
 | |
3CDT
 
 | |
3CDV
 
 | |
3C80
 
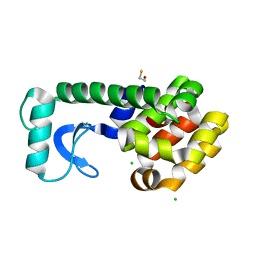 | | T4 Lysozyme mutant R96Y at room temperature | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3C8R
 
 | |
3CDO
 
 | |
