1Q18
 
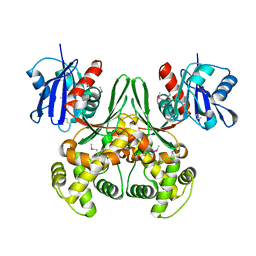 | | Crystal structure of E.coli glucokinase (Glk) | | Descriptor: | Glucokinase | | Authors: | Lunin, V.V, Li, Y, Schrag, J.D, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-07-18 | | Release date: | 2004-07-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose.
J.Bacteriol., 186, 2004
|
|
1PTM
 
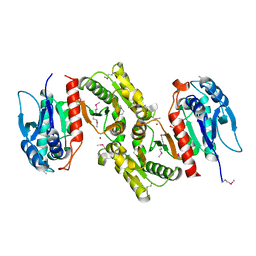 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, PHOSPHATE ION, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-06-23 | | Release date: | 2003-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
4MLM
 
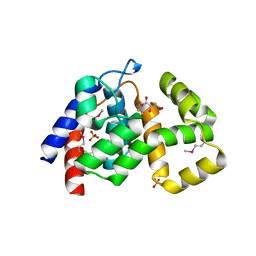 | | Crystal Structure of PhnZ from uncultured bacterium HF130_AEPn_1 | | Descriptor: | FE (III) ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | van Staalduinen, L.M, McSorley, F.R, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of PhnZ in complex with substrate reveals a di-iron oxygenase mechanism for catabolism of organophosphonates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MLN
 
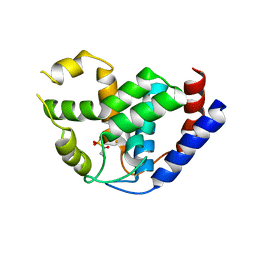 | | Crystal of PhnZ bound to (R)-2-amino-1-hydroxyethylphosphonic acid | | Descriptor: | FE (III) ION, Predicted HD phosphohydrolase PhnZ, [(1R)-2-amino-1-hydroxyethyl]phosphonic acid | | Authors: | van Staalduinen, L.M, McSorley, F.R, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PhnZ in complex with substrate reveals a di-iron oxygenase mechanism for catabolism of organophosphonates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3G2Q
 
 | | Crystal Structure of the Glycopeptide N-methyltransferase MtfA complexed with sinefungin | | Descriptor: | PCZA361.24, SINEFUNGIN | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics.
Chem.Biol., 16, 2009
|
|
3G2P
 
 | | Crystal Structure of the Glycopeptide N-methyltransferase MtfA complexed with (S)-adenosyl-L-homocysteine (SAH) | | Descriptor: | PCZA361.24, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-31 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics.
Chem.Biol., 16, 2009
|
|
3G05
 
 | | Crystal structure of N-terminal domain (2-550) of E.coli MnmG | | Descriptor: | SULFATE ION, tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-27 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme.
J.Bacteriol., 191, 2009
|
|
5K35
 
 | | Structure of the Legionella effector, AnkB, in complex with human Skp1 | | Descriptor: | Ankyrin-repeat protein B, S-phase kinase-associated protein 1 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
5K34
 
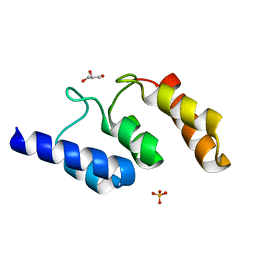 | | Structure of the ankyrin domain of AnkB from Legionella Pneumophila | | Descriptor: | Ankyrin-repeat protein B, GLYCEROL, SULFATE ION | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-05-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Mimicry by a Bacterial F Box Effector Hijacks the Host Ubiquitin-Proteasome System.
Structure, 25, 2017
|
|
5CQ9
 
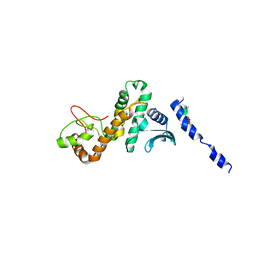 | |
5KDG
 
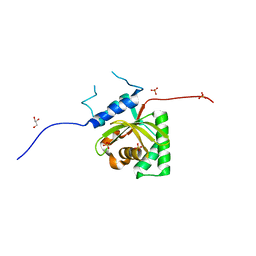 | | Crystal Structure of Salmonella Typhimurium Effector GtgE | | Descriptor: | GLYCEROL, Gifsy-2 prophage protein, SULFATE ION | | Authors: | Kozlov, G, Xu, C, Wong, K, Gehring, K, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2016-06-08 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the Salmonella Typhimurium Effector GtgE.
PLoS ONE, 11, 2016
|
|
5VRQ
 
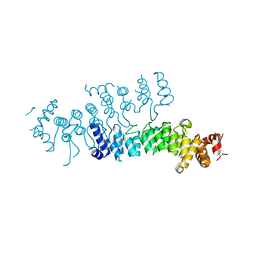 | | Crystal structure of Legionella pneumophila effector AnkC | | Descriptor: | Ankyrin repeat-containing protein | | Authors: | Kozlov, G, Wong, K, Wang, W, Skubak, P, Munoz-Escobar, J, Liu, Y, Pannu, N.S, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2017-05-11 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Ankyrin repeats as a dimerization module.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5WD8
 
 | |
5WD9
 
 | |
1KSL
 
 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URACIL | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-01-13 | | Release date: | 2002-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1K75
 
 | | The L-histidinol dehydrogenase (hisD) structure implicates domain swapping and gene duplication. | | Descriptor: | GLYCEROL, L-histidinol dehydrogenase, SULFATE ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-10-18 | | Release date: | 2002-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KK9
 
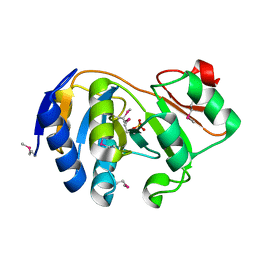 | | CRYSTAL STRUCTURE OF E. COLI YCIO | | Descriptor: | SULFATE ION, probable translation factor yciO | | Authors: | Jia, J, Lunin, V.V, Sauve, V, Huang, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-12-06 | | Release date: | 2002-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the YciO protein from Escherichia coli
PROTEINS: STRUCT.,FUNCT.,GENET., 49, 2002
|
|
1LKZ
 
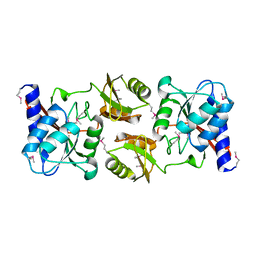 | | Crystal structure of D-ribose-5-phosphate isomerase (RpiA) from Escherichia coli. | | Descriptor: | Ribose 5-phosphate isomerase A | | Authors: | Rangarajan, E.S, Sivaraman, J, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-04-26 | | Release date: | 2002-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of D-ribose-5-phosphate isomerase (RpiA) from Escherichia coli
Proteins, 48, 2002
|
|
1KON
 
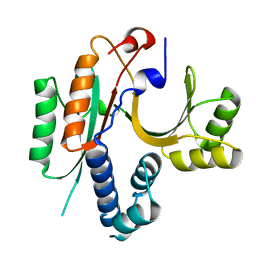 | | CRYSTAL STRUCTURE OF E.COLI YEBC | | Descriptor: | Protein yebC | | Authors: | Jia, J, Smith, C, Lunin, V.V, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-12-21 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | UNPUBLISHED
TO BE PUBLISHED
|
|
3UCS
 
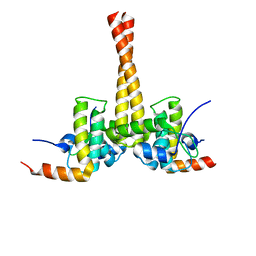 | |
5BTW
 
 | | Structure of the N-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
3GZH
 
 | |
1FG7
 
 | |
1FC5
 
 | |
5BTZ
 
 | | Structure of the middle domain of lpg1496 from Legionella pneumophila in P212121 space group | | Descriptor: | lpg1496 | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
