7M5T
 
 | |
6UN9
 
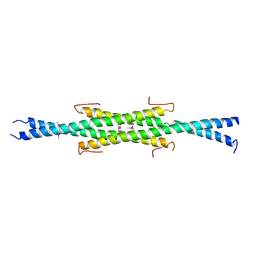 | | Crystal Structure of the Q7VLF5_HAEDU protein from Haemophilus ducreyi. Northeast Structural Genomics Consortium Target Hdr25 | | Descriptor: | Uncharacterized protein | | Authors: | Vorobiev, S.M, Seetharaman, J, Kolev, M, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2019-10-11 | | Release date: | 2020-12-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Q7VLF5_HAEDU protein from Haemophilus ducreyi. Northeast Structural Genomics Consortium Target Hdr25
To Be Published
|
|
1IHQ
 
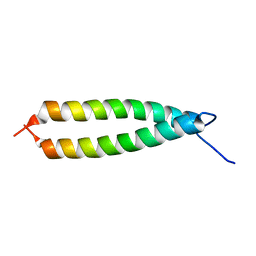 | | GLYTM1BZIP: A CHIMERIC PEPTIDE MODEL OF THE N-TERMINUS OF A RAT SHORT ALPHA TROPOMYOSIN WITH THE N-TERMINUS ENCODED BY EXON 1B | | Descriptor: | CHIMERIC PEPTIDE GlyTM1bZip: TROPOMYOSIN ALPHA CHAIN, BRAIN-3 and GENERAL CONTROL PROTEIN GCN4 | | Authors: | Greenfield, N.J, Yuang, Y.J, Palm, T, Swapna, G.V, Monleon, D, Montelione, G.T, Hitchcock-Degregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and folding dynamics of the N terminus of a rat non-muscle alpha-tropomyosin in an engineered chimeric protein.
J.Mol.Biol., 312, 2001
|
|
7JMY
 
 | |
7JYZ
 
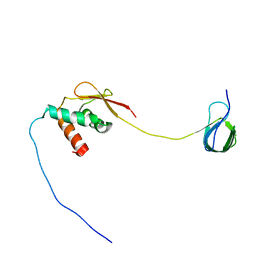 | | Solution NMR structure and dynamics of human Brd3 ET in complex with MLV IN CTD | | Descriptor: | Bromodomain-containing protein 3, Integrase | | Authors: | Aiyer, S, Liu, G, Swapna, G.V.T, Hao, J, Ma, L.C, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-09-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
7JYN
 
 | | Solution NMR structure of human Brd3 ET complexed with NSD3(148-184) peptide | | Descriptor: | Bromodomain-containing protein 3, Histone-lysine N-methyltransferase NSD3 | | Authors: | Aiyer, S, Swapna, G.V.T, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
4PWW
 
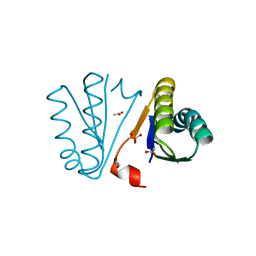 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR494. | | Descriptor: | ACETIC ACID, OR494, PHOSPHATE ION | | Authors: | Vorobiev, S, Lin, Y.-R, Seetharaman, J, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Crystal Structure of Engineered Protein OR494.
To be Published
|
|
4R80
 
 | | Crystal Structure of a De Novo Designed Beta Sheet Protein, Cystatin Fold, Northeast Structural Genomics Consortium (NESG) Target OR486 | | Descriptor: | OR486 | | Authors: | Guan, R, Marcos, E, O'Connell, P, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Crystal Structure of an engineered protein with denovo beta sheet design, Northeast Structural Genomics Consortium (NESG) Target OR486
To be Published
|
|
4RJV
 
 | | Crystal Structure of a De Novo Designed Ferredoxin Fold, Northeast Structural Genomics Consortium (NESG) Target OR461 | | Descriptor: | OR461 | | Authors: | O'Connell, P.T, Lin, Y.-R, Guan, R, Koga, N, Koga, R, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR461
To be published
|
|
4RRP
 
 | | Crystal Structure of the Fab complexed with antigen Asf1p, Northeast Structural Genomics Consortium (NESG) Target PdR16 | | Descriptor: | Antigen Asf1p, DI(HYDROXYETHYL)ETHER, Fab antibody, ... | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Mao, L, Xiao, R, Oconnell, P.T, Maglaqui, M, Bailey, L, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structure of the Fab complexed with antigen Asf1p, Northeast Structural Genomics Consortium (NESG) Target PdR16
To be Published
|
|
4RZP
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium (NESG) Target OR366. | | Descriptor: | Engineered Protein OR366 | | Authors: | Vorobiev, S, Parmeggiani, F, Dimaio, F, Seetharaman, J, Sahdev, S, Xiao, R, Kogan, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-12-23 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Crystal Structure of Engineered Protein OR366
To be Published
|
|
4RMM
 
 | | Crystal Structure of the Q7NVP2_CHRVO protein from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvR191 | | Descriptor: | Putative uncharacterized protein | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Mao, L, Xiao, R, Ciccosanti, C, Foote, E.L, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-10-21 | | Release date: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Q7NVP2_CHRVO protein from Chromobacterium violaceum.
To be Published
|
|
4RV1
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium (NESG) Target OR497. | | Descriptor: | ACETATE ION, Engineered Protein OR497 | | Authors: | Vorobiev, S, Parmeggiani, F, Seetharaman, J, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-11-24 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Crystal Structure of Engineered Protein OR497.
To be Published
|
|
4RL6
 
 | | Crystal Structure of the Q04L03_STRP2 protein from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR105 | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Saccharopine dehydrogenase | | Authors: | Vorobiev, S, Neely, H.M, Odukwe, C.D, Seetharaman, J, Mao, L, Xiao, R, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structure of the Q04L03_STRP2 protein from Streptococcus pneumoniae.
To be Published
|
|
6NS8
 
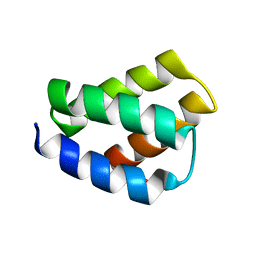 | | RDC-refined SOLUTION NMR STRUCTURE OF PROTEIN PF2048.1 | | Descriptor: | Uncharacterized protein | | Authors: | Daigham, N.S, Liu, G, Swapna, G.V.T, Cole, C, Valafar, H, Montelione, G.T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | REDCRAFT: A Computational Platform Using Residual Dipolar Coupling NMR Data for Determining Structures of Perdeuterated Proteins Without NOEs
To Be Published
|
|
5KVJ
 
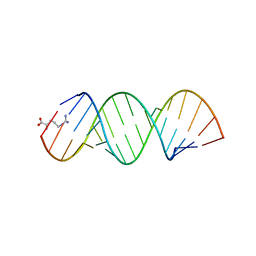 | |
6E4H
 
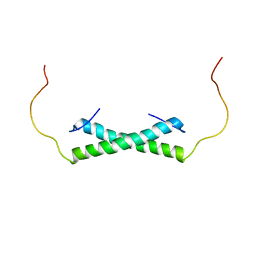 | | Solution NMR Structure of the Colied-coil PALB2 Homodimer | | Descriptor: | Partner and localizer of BRCA2 | | Authors: | Song, F, Li, M, Liu, G, Swapna, G.V.T, Xia, B, Bunting, S.F, Montelione, G.T. | | Deposit date: | 2018-07-17 | | Release date: | 2018-10-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Antiparallel Coiled-Coil Interactions Mediate the Homodimerization of the DNA Damage-Repair Protein PALB2.
Biochemistry, 57, 2018
|
|
6E4J
 
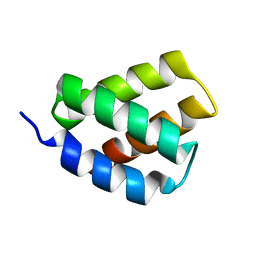 | | Solution NMR Structure of Protein PF2048.1 | | Descriptor: | Uncharacterized protein PF2048.1 | | Authors: | Daigham, N.S, Liu, G, Swapna, G.V.T, Cole, C, Valafar, H, Montelione, G.T. | | Deposit date: | 2018-07-17 | | Release date: | 2018-08-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | REDCRAFT: A Computational Platform Using Residual Dipolar Coupling NMR Data for Determining Structures of Perdeuterated Proteins Without NOEs
To Be Published
|
|
1IHN
 
 | |
1L7Y
 
 | | Solution NMR Structure of C. elegans Protein ZK652.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR41. | | Descriptor: | HYPOTHETICAL PROTEIN ZK652.3 | | Authors: | Cort, J.R, Chiang, Y, Zheng, D, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-18 | | Release date: | 2002-08-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of conserved eukaryotic protein ZK652.3 from C. elegans: a ubiquitin-like fold.
Proteins, 48, 2002
|
|
1L7B
 
 | | Solution NMR Structure of BRCT Domain of T. Thermophilus: Northeast Structural Genomics Consortium Target WR64TT | | Descriptor: | DNA LIGASE | | Authors: | Sahota, G, Dixon, B.L, Huang, Y.P, Aramini, J, Monleon, D, Bhattacharya, D, Swapna, G.V.T, Yin, C, Xiao, R, Anderson, S, Tejero, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-14 | | Release date: | 2003-09-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Brct Domain from Thermus Thermophilus DNA Ligase
To be Published
|
|
1Q2N
 
 | |
7TZ8
 
 | |
7TZD
 
 | |
7UBI
 
 | |
