3IR4
 
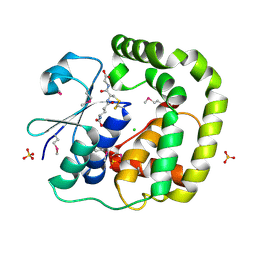 | | 1.2 Angstrom Crystal Structure of the Glutaredoxin 2 (grxB) from Salmonella typhimurium in complex with Glutathione | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutaredoxin 2, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Halavaty, A, Dauter, Z, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-01 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 1.2 Angstrom Crystal Structure of the Glutaredoxin 2 (grxB) from Salmonella typhimurium in complex with Glutathione.
TO BE PUBLISHED
|
|
4HS7
 
 | | 2.6 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with PEG. | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Bacterial extracellular solute-binding protein, putative, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-10-29 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with PEG.
TO BE PUBLISHED
|
|
3IJ5
 
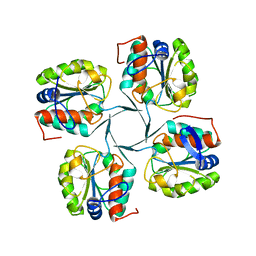 | | 1.95 Angstrom Resolution Crystal Structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Yersinia pestis | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CHLORIDE ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-03 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Yersinia pestis
TO BE PUBLISHED
|
|
3INP
 
 | | 2.05 Angstrom Resolution Crystal Structure of D-ribulose-phosphate 3-epimerase from Francisella tularensis. | | Descriptor: | CHLORIDE ION, D-ribulose-phosphate 3-epimerase, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Scott, P, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-12 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Resolution Crystal Structure of D-ribulose-phosphate 3-epimerase from Francisella tularensis.
TO BE PUBLISHED
|
|
3IAH
 
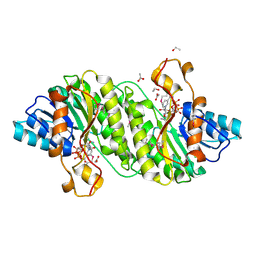 | | Crystal Structure of Short Chain Dehydrogenase (yciK) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 in Complex with NADP and Acetate. | | Descriptor: | ACETATE ION, CHLORIDE ION, ETHANOL, ... | | Authors: | Minasov, G, Halavaty, A, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-14 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Short Chain Dehydrogenase (yciK) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 in Complex with NADP and Acetate.
TO BE PUBLISHED
|
|
3IGX
 
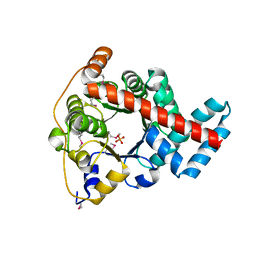 | | 1.85 Angstrom Resolution Crystal Structure of Transaldolase B (talA) from Francisella tularensis. | | Descriptor: | PHOSPHATE ION, Transaldolase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Gordon, E, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-11 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Transaldolase B (talA) from Francisella tularensis.
TO BE PUBLISHED
|
|
7KOM
 
 | | High Resolution Crystal Structure of Putative Pterin Binding Protein PruR (VV2_1280) from Vibrio vulnificus CMCP6 | | Descriptor: | FORMIC ACID, Oxidored_molyb domain-containing protein, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High Resolution Crystal Structure of Putative Pterin Binding Protein PruR (VV2_1280) from Vibrio vulnificus CMCP6.
To Be Published
|
|
7KOU
 
 | | 1.83 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Pterin Binding Protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-10 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58.
To Be Published
|
|
7KP2
 
 | | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin | | Descriptor: | L-NEOPTERIN, Putative Pterin Binding Protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Pshenychnyi, S, Dubrovska, I, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-10 | | Release date: | 2021-11-17 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin.
To Be Published
|
|
7KOS
 
 | | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58 | | Descriptor: | FORMIC ACID, MALONIC ACID, Pterin Binding Protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58.
To Be Published
|
|
4HW8
 
 | | 2.25 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with Maltose. | | Descriptor: | Bacterial extracellular solute-binding protein, putative, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-07 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | 2.25 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with Maltose.
TO BE PUBLISHED
|
|
7L6R
 
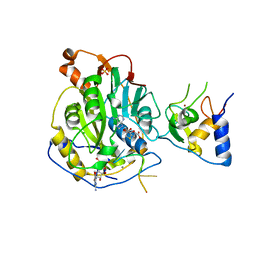 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA2m)pUpUpApApA (Cap-1), S-Adenosyl-L-homocysteine (SAH) and Manganese (Mn). | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mn 2+ coordinates Cap-0-RNA to align substrates for efficient 2'- O -methyl transfer by SARS-CoV-2 nsp16.
Sci.Signal., 14, 2021
|
|
7L6T
 
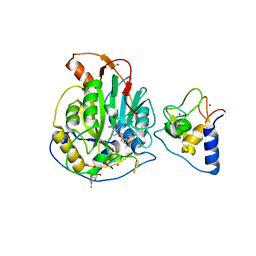 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA2m)pUpUpApApA (Cap-1), S-Adenosyl-L-homocysteine (SAH) and two Magnesium (Mg) ions. | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-23 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mn 2+ coordinates Cap-0-RNA to align substrates for efficient 2'- O -methyl transfer by SARS-CoV-2 nsp16.
Sci.Signal., 14, 2021
|
|
3JZE
 
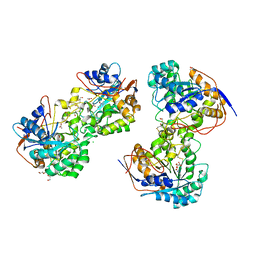 | | 1.8 Angstrom resolution crystal structure of dihydroorotase (pyrC) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | ACETIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Dihydroorotase (pyrC) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2.
TO BE PUBLISHED
|
|
7L75
 
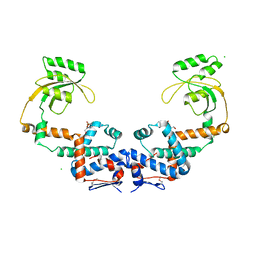 | | Crystal Structure of Peptidylprolyl Isomerase PrsA from Streptococcus mutans. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Foldase protein PrsA | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-25 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure of Peptidylprolyl Isomerase PrsA from Streptococcus mutans.
To Be Published
|
|
7L6Z
 
 | | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-24 | | Release date: | 2021-12-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6
To Be Published
|
|
7L6Y
 
 | |
7L71
 
 | |
3K96
 
 | | 2.1 Angstrom resolution crystal structure of glycerol-3-phosphate dehydrogenase (gpsA) from Coxiella burnetii | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, Glycerol-3-phosphate dehydrogenase [NAD(P)+] | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Glycerol-3-phosphate Dehydrogenase (gpsA) from Coxiella burnetii.
TO BE PUBLISHED
|
|
7L5T
 
 | | Crystal Structure of the Oxacillin-hydrolyzing Class D Extended-spectrum Beta-lactamase OXA-14 from Pseudomonas aeruginosa in Complex with Covalently Bound Clavulanic Acid | | Descriptor: | (2E)-3-[(4-hydroxy-2-oxobutyl)amino]prop-2-enal, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Membrane Proteins of Infectious Diseases (MPID) | | Deposit date: | 2020-12-22 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of the Oxacillin-hydrolyzing Class D Extended-spectrum Beta-lactamase OXA-14 from Pseudomonas aeruginosa in Complex with Covalently Bound Clavulanic Acid
To Be Published
|
|
3KBO
 
 | | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Glyoxylate/hydroxypyruvate reductase A, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | 2.14 Angstrom Crystal Structure of Putative Oxidoreductase (ycdW) from Salmonella typhimurium in Complex with NADP.
To be Published
|
|
3KQF
 
 | | 1.8 Angstrom Resolution Crystal Structure of Enoyl-CoA Hydratase from Bacillus anthracis. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Minasov, G, Halavaty, A, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-17 | | Release date: | 2009-11-24 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Enoyl-CoA Hydratase from Bacillus anthracis.
TO BE PUBLISHED
|
|
3LB0
 
 | | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Citrate Bound to the Active Site. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinate dehydratase, CITRIC ACID, ... | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-07 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Citrate Bound to the Active Site.
TO BE PUBLISHED
|
|
3R2U
 
 | | 2.1 Angstrom Resolution Crystal Structure of Metallo-beta-lactamase from Staphylococcus aureus subsp. aureus COL | | Descriptor: | CHLORIDE ION, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-14 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Metallo-beta-lactamase Family Protein from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
3RPE
 
 | | 1.1 Angstrom Crystal Structure of Putative Modulator of Drug Activity (MdaB) from Yersinia pestis CO92. | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, Modulator of drug activity B | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-26 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | 1.1 Angstrom Crystal Structure of Putative Modulator of Drug Activity (MdaB) from Yersinia pestis CO92.
TO BE PUBLISHED
|
|
