1IUA
 
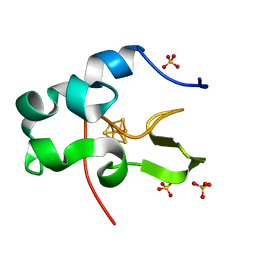 | | Ultra-high resolution structure of HiPIP from Thermochromatium tepidum | | Descriptor: | High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Liu, L, Nogi, T, Kobayashi, M, Nozawa, T, Miki, K. | | Deposit date: | 2002-03-01 | | Release date: | 2002-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Ultrahigh-resolution structure of high-potential iron-sulfur protein from Thermochromatium tepidum.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GEH
 
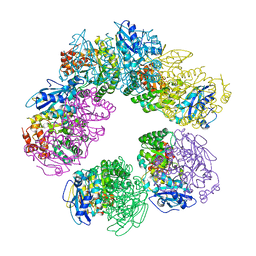 | | CRYSTAL STRUCTURE OF ARCHAEAL RUBISCO (RIBULOSE 1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE) | | Descriptor: | RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE, SULFATE ION | | Authors: | Kitano, K, Maeda, N, Fukui, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2000-11-13 | | Release date: | 2001-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Novel-Type Archaeal Rubisco with Pentagonal Symmetry
Structure, 9, 2001
|
|
1X0P
 
 | | Structure of a cyanobacterial BLUF protein, Tll0078 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, hypothetical protein Tll0078 | | Authors: | Kita, A, Okajima, K, Morimoto, Y, Ikeuchi, M, Miki, K. | | Deposit date: | 2005-03-27 | | Release date: | 2005-06-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Cyanobacterial BLUF Protein, Tll0078, Containing a Novel FAD-binding Blue Light Sensor Domain
J.Mol.Biol., 349, 2005
|
|
2D05
 
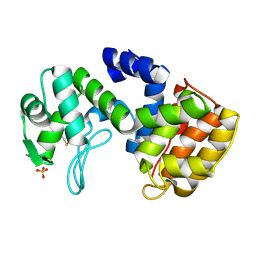 | | Chitosanase From Bacillus circulans mutant K218P | | Descriptor: | Chitosanase, SULFATE ION | | Authors: | Fukamizo, T, Amano, S, Yamaguchi, K, Yoshikawa, T, Katsumi, T, Saito, J, Suzuki, M, Miki, K, Nagata, Y, Ando, A. | | Deposit date: | 2005-07-25 | | Release date: | 2005-12-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus circulans MH-K1 Chitosanase: Amino Acid Residues Responsible for Substrate Binding
J.Biochem.(Tokyo), 138, 2005
|
|
1OWM
 
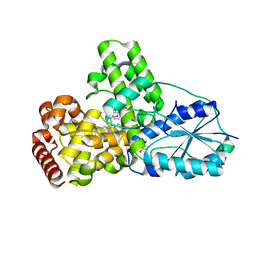 | | DATA1:DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWN
 
 | | DATA3:DNA photolyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWP
 
 | | DATA6:photoreduced DNA pholyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWL
 
 | | Structure of apophotolyase from Anacystis nidulans | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7E4L
 
 | |
7E96
 
 | | Oxy-deoxy intermediate of 400 kDa giant hemoglobin at 69% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular giant hemoglobin major globin subunit A1, Extracellular giant hemoglobin major globin subunit A2, ... | | Authors: | Numoto, N, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Coarse snapshots of oxygen-dissociation intermediates of a giant hemoglobin elucidated by determining the oxygen saturation in individual subunits in the crystalline state.
Iucrj, 8, 2021
|
|
7E99
 
 | | Oxy-deoxy intermediate of 400 kDa giant hemoglobin at 13% oxygen saturation | | Descriptor: | Extracellular giant hemoglobin major globin subunit A1, Extracellular giant hemoglobin major globin subunit A2, Extracellular giant hemoglobin major globin subunit B2, ... | | Authors: | Numoto, N, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Coarse snapshots of oxygen-dissociation intermediates of a giant hemoglobin elucidated by determining the oxygen saturation in individual subunits in the crystalline state.
Iucrj, 8, 2021
|
|
7E97
 
 | | Oxy-deoxy intermediate of 400 kDa giant hemoglobin at 58% oxygen saturation | | Descriptor: | Extracellular giant hemoglobin major globin subunit A1, Extracellular giant hemoglobin major globin subunit A2, Extracellular giant hemoglobin major globin subunit B2, ... | | Authors: | Numoto, N, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Coarse snapshots of oxygen-dissociation intermediates of a giant hemoglobin elucidated by determining the oxygen saturation in individual subunits in the crystalline state.
Iucrj, 8, 2021
|
|
7E98
 
 | | Oxy-deoxy intermediate of 400 kDa giant hemoglobin at 21% oxygen saturation | | Descriptor: | Extracellular giant hemoglobin major globin subunit A1, Extracellular giant hemoglobin major globin subunit A2, Extracellular giant hemoglobin major globin subunit B2, ... | | Authors: | Numoto, N, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Coarse snapshots of oxygen-dissociation intermediates of a giant hemoglobin elucidated by determining the oxygen saturation in individual subunits in the crystalline state.
Iucrj, 8, 2021
|
|
6JBC
 
 | | Phosphotransferase related to CoA biosynthesis pathway | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Kita, A, Kishimoto, A, Shimosaka, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pantoate kinase from Thermococcus kodakarensis.
Proteins, 88, 2020
|
|
6JBD
 
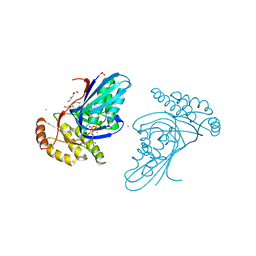 | | Phosphotransferase-ATP complex related to CoA biosynthesis pathway | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kita, A, Kishimoto, A, Shimosaka, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of pantoate kinase from Thermococcus kodakarensis.
Proteins, 88, 2020
|
|
1EYS
 
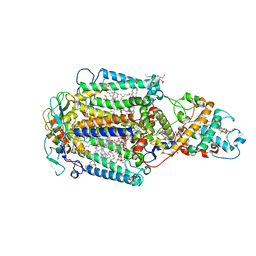 | | CRYSTAL STRUCTURE OF PHOTOSYNTHETIC REACTION CENTER FROM A THERMOPHILIC BACTERIUM, THERMOCHROMATIUM TEPIDUM | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Nogi, T, Fathir, I, Kobayashi, M, Nozawa, T, Miki, K. | | Deposit date: | 2000-05-08 | | Release date: | 2000-12-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of photosynthetic reaction center and high-potential iron-sulfur protein from Thermochromatium tepidum: thermostability and electron transfer.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5B3Q
 
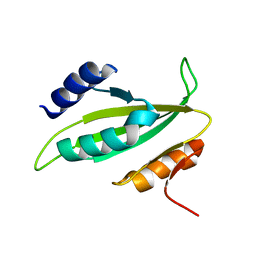 | |
5B3Y
 
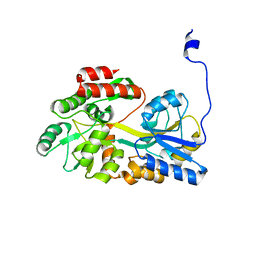 | |
5B3W
 
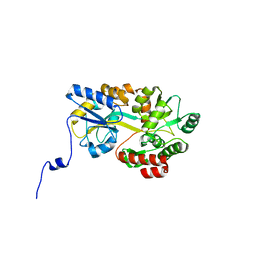 | | Crystal structure of hPin1 WW domain (5-15) fused with maltose-binding protein in C2221 form | | Descriptor: | CITRIC ACID, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1,Maltose-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hanazono, Y, Takeda, K, Miki, K. | | Deposit date: | 2016-03-17 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of the N-terminal fragments of the WW domain: Insights into co-translational folding of a beta-sheet protein
Sci Rep, 6, 2016
|
|
5B3X
 
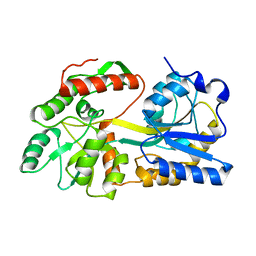 | |
5B3Z
 
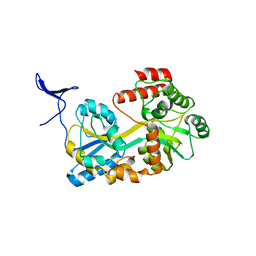 | |
5B3P
 
 | |
5BMY
 
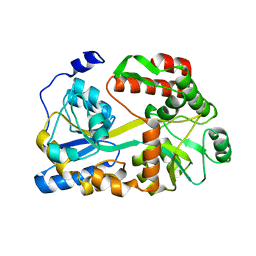 | |
5D8V
 
 | | Ultra-high resolution structure of high-potential iron-sulfur protein | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Hirano, Y, Takeda, K, Miki, K. | | Deposit date: | 2015-08-18 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.48 Å) | | Cite: | Charge-density analysis of an iron-sulfur protein at an ultra-high resolution of 0.48 angstrom
Nature, 534, 2016
|
|
5DHE
 
 | | Crystal structure of ChBD3 from Thermococcus kodakarensis KOD1 | | Descriptor: | Chitinase, GLYCEROL | | Authors: | Niwa, S, Hibi, M, Takeda, K, Miki, K. | | Deposit date: | 2015-08-30 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of chitin binding domains of chitinase from Thermococcus kodakarensis KOD1
Febs Lett., 590, 2016
|
|
