3FEV
 
 | | Crystal structure of the chimeric muscarinic toxin MT7 with loop 1 from MT1. | | Descriptor: | Fusion of Muscarinic toxin 1, Muscarinic m1-toxin1, SULFATE ION | | Authors: | Stura, E.A, Menez, R, Mourier, G, Fruchart-Gaillard, C, Menez, A, Servant, D. | | Deposit date: | 2008-12-01 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering of three-finger fold toxins creates ligands with original pharmacological profiles for muscarinic and adrenergic receptors.
Plos One, 7, 2012
|
|
2H7Z
 
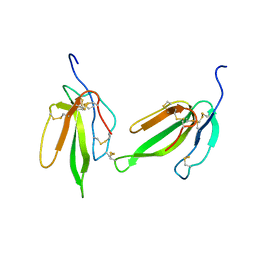 | | Crystal structure of irditoxin | | Descriptor: | Irditoxin subunit A, Irditoxin subunit B | | Authors: | Pawlak, J, Kini, R.M, Stura, E.A, Le Du, M.H. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-29 | | Last modified: | 2011-10-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity.
Faseb J., 23, 2009
|
|
2H5F
 
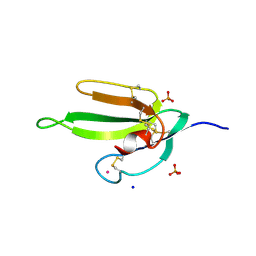 | | Denmotoxin: A the three-finger toxin from colubrid snake Boiga dendrophila with bird-specific activity | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Pawlak, J, Kini, R.M, Stura, E.A. | | Deposit date: | 2006-05-26 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Denmotoxin, a three-finger toxin from the colubrid snake Boiga dendrophila (Mangrove Catsnake) with bird-specific activity.
J.Biol.Chem., 281, 2006
|
|
3MK1
 
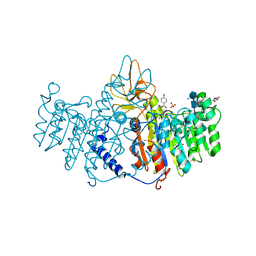 | | Refinement of placental alkaline phosphatase complexed with nitrophenyl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Alkaline phosphatase, ... | | Authors: | Stec, B, Cheltsov, A, Millan, J.L. | | Deposit date: | 2010-04-13 | | Release date: | 2011-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Refined structures of placental alkaline phosphatase show a consistent pattern of interactions at the peripheral site.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3MK0
 
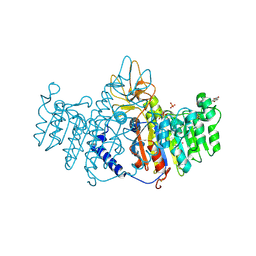 | | Refinement of placental alkaline phosphatase complexed with nitrophenyl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Alkaline phosphatase, ... | | Authors: | Stec, B, Cheltsov, A, Millan, J.L. | | Deposit date: | 2010-04-13 | | Release date: | 2011-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined structures of placental alkaline phosphatase show a consistent pattern of interactions at the peripheral site.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3MK2
 
 | | Placental alkaline phosphatase complexed with Phe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Alkaline phosphatase, ... | | Authors: | Stec, B, Cheltsov, A, Millan, J.L. | | Deposit date: | 2010-04-13 | | Release date: | 2011-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Refined structures of placental alkaline phosphatase show a consistent pattern of interactions at the peripheral site.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2MOA
 
 | |
1KH9
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153GD330N) COMPLEX WITH PHOSPHATE | | Descriptor: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KH7
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153GD330N) | | Descriptor: | MAGNESIUM ION, SULFATE ION, ZINC ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KH5
 
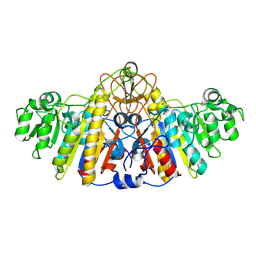 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D330N) MIMIC OF THE TRANSITION STATES WITH ALUMINIUM FLUORIDE | | Descriptor: | ALKALINE PHOSPHATASE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KH4
 
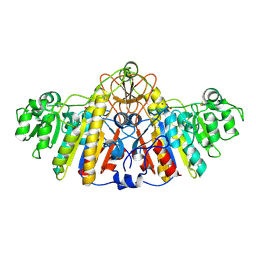 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D330N) IN COMPLEX WITH PHOSPHATE | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1K7H
 
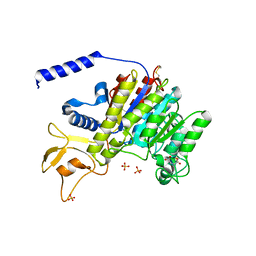 | | CRYSTAL STRUCTURE OF SHRIMP ALKALINE PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALKALINE PHOSPHATASE, MALEIC ACID, ... | | Authors: | De Backer, M.E, Mc Sweeney, S, Rasmussen, H.B, Riise, B.W, Lindley, P, Hough, E. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The 1.9 A Crystal Structure of Heat-Labile Shrimp Alkaline Phosphatase
J.Mol.Biol., 318, 2002
|
|
1M6J
 
 | |
8P4Q
 
 | | Structure of the IMP dehydrogenase related protein GUAB3 from Synechocystis PCC 6803 | | Descriptor: | IMP dehydrogenase subunit, INOSINIC ACID, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Hernandez-Gomez, A, Fernandez-Justel, D, Buey, R.M. | | Deposit date: | 2023-05-23 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | GuaB3, an overlooked enzyme in cyanobacteria's toolbox that sheds light on IMP dehydrogenase evolution.
Structure, 31, 2023
|
|
8P37
 
 | | Structure a catalytically inactive mutant of the IMP dehydrogenase related protein GUAB3 from Synechocystis PCC 6803 | | Descriptor: | IMP dehydrogenase subunit, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Hernandez-Gomez, A, Fernandez-Justel, D, Buey, R.M. | | Deposit date: | 2023-05-17 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | GuaB3, an overlooked enzyme in cyanobacteria's toolbox that sheds light on IMP dehydrogenase evolution.
Structure, 31, 2023
|
|
7PJI
 
 | |
6VCB
 
 | | Cryo-EM structure of the Glucagon-like peptide-1 receptor in complex with G protein, GLP-1 peptide and a positive allosteric modulator | | Descriptor: | 1-[(1R)-1-(2,6-dichloro-3-methoxyphenyl)ethyl]-6-{2-[(2R)-piperidin-2-yl]phenyl}-1H-benzimidazole, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Sun, B, Feng, D, Bueno, A, Kobilka, B, Sloop, K. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into probe-dependent positive allosterism of the GLP-1 receptor.
Nat.Chem.Biol., 16, 2020
|
|
7PMZ
 
 | | Crystal structure of Streptomyces coelicolor guaB (IMP dehydrogenase) bound to ATP and ppGpp at 2.0 A resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5',3'-TETRAPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Fernandez-Justel, D, Revuelta, J.L, Buey, R.M. | | Deposit date: | 2021-09-04 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Diversity of mechanisms to control bacterial GTP homeostasis by the mutually exclusive binding of adenine and guanine nucleotides to IMP dehydrogenase.
Protein Sci., 31, 2022
|
|
