1L98
 
 | | PERTURBATION OF TRP 138 IN T4 LYSOZYME BY MUTATIONS AT GLN 105 USED TO CORRELATE CHANGES IN STRUCTURE, STABILITY, SOLVATION, AND SPECTROSCOPIC PROPERTIES | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Pjura, P, Mcintosh, L.P, Wozniak, J.A, Matthews, B.W. | | Deposit date: | 1992-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Perturbation of Trp 138 in T4 lysozyme by mutations at Gln 105 used to correlate changes in structure, stability, solvation, and spectroscopic properties.
Proteins, 15, 1993
|
|
1L00
 
 | | PERTURBATION OF TRP 138 IN T4 LYSOZYME BY MUTATIONS AT GLN 105 USED TO CORRELATE CHANGES IN STRUCTURE, STABILITY, SOLVATION, AND SPECTROSCOPIC PROPERTIES | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Pjura, P, Mcintosh, L.P, Wozniak, J.A, Matthews, B.W. | | Deposit date: | 1992-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Perturbation of Trp 138 in T4 lysozyme by mutations at Gln 105 used to correlate changes in structure, stability, solvation, and spectroscopic properties.
Proteins, 15, 1993
|
|
4W4M
 
 | | Crystal structure of PrgK 19-92 | | Descriptor: | Lipoprotein PrgK | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2014-08-15 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Modular Structure of the Inner-Membrane Ring Component PrgK Facilitates Assembly of the Type III Secretion System Basal Body.
Structure, 23, 2015
|
|
3J6D
 
 | | Model of the PrgH-PrgK periplasmic rings | | Descriptor: | Pathogenicity 1 island effector protein, Protein PrgH | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2014-02-14 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | The Modular Structure of the Inner-Membrane Ring Component PrgK Facilitates Assembly of the Type III Secretion System Basal Body.
Structure, 23, 2015
|
|
6CCD
 
 | |
1BVV
 
 | |
2MKY
 
 | |
5ILS
 
 | | Autoinhibited ETV1 | | Descriptor: | ETS translocation variant 1 | | Authors: | Whitby, F.G, Currie, S.L. | | Deposit date: | 2016-03-04 | | Release date: | 2017-02-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETS factors ETV1, ETV4 and ETV5.
Nucleic Acids Res., 45, 2017
|
|
5ILV
 
 | | Uninhibited ETV5 | | Descriptor: | ETS translocation variant 5 | | Authors: | Whitby, F.G, Currie, S.L. | | Deposit date: | 2016-03-04 | | Release date: | 2017-02-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETS factors ETV1, ETV4 and ETV5.
Nucleic Acids Res., 45, 2017
|
|
5ILU
 
 | | Autoinhibited ETV4 | | Descriptor: | ETS translocation variant 4 | | Authors: | Whitby, F.G, Currie, S.L. | | Deposit date: | 2016-03-04 | | Release date: | 2017-02-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETS factors ETV1, ETV4 and ETV5.
Nucleic Acids Res., 45, 2017
|
|
6UM9
 
 | |
6XJ6
 
 | |
6XJ7
 
 | |
2JV3
 
 | |
2KXX
 
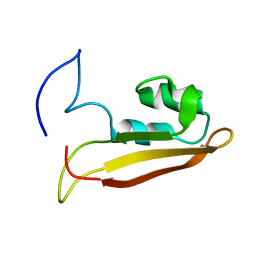 | | NMR Structure of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex | | Descriptor: | Small protein A | | Authors: | Kim, K, Okon, M, Escobar, E, Kang, H, McIntosh, L, Paetzel, M. | | Deposit date: | 2010-05-13 | | Release date: | 2011-01-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex.
Biochemistry, 50, 2011
|
|
1BSH
 
 | |
1BSN
 
 | |
3CUI
 
 | |
3CUG
 
 | |
3CUH
 
 | |
3CUJ
 
 | |
3CUF
 
 | |
3ECH
 
 | |
2BVV
 
 | |
2Q2I
 
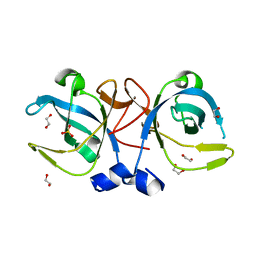 | | Crystal structure of the protein secretion chaperone CsaA from Agrobacterium tumefaciens. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Secretion chaperone | | Authors: | Feldman, A.R, Shapova, Y.A, Paetzel, M. | | Deposit date: | 2007-05-28 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Phage display and crystallographic analysis reveals potential substrate/binding site interactions in the protein secretion chaperone CsaA from Agrobacterium tumefaciens.
J.Mol.Biol., 379, 2008
|
|
