7Q2A
 
 | | Crystal structure of AphC in complex with 4-ethylcatechol | | Descriptor: | 4-ethylbenzene-1,2-diol, CALCIUM ION, Catechol 2,3-dioxygenase, ... | | Authors: | Zahn, M, Grigg, J.C, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2021-10-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization of a phylogenetically distinct extradiol dioxygenase involved in the bacterial catabolism of lignin-derived aromatic compounds.
J.Biol.Chem., 298, 2022
|
|
7QWT
 
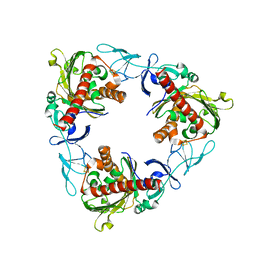 | | Rieske non-heme iron monooxygenase for guaiacol O-demethylation | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Rieske (2Fe-2S) domain protein | | Authors: | Hinchen, D.J, Zahn, M, Bleem, A, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (3.712 Å) | | Cite: | Discovery, characterization, and metabolic engineering of Rieske non-heme iron monooxygenases for guaiacol O-demethylation
Chem Catal, 2022
|
|
8AYV
 
 | |
4X4F
 
 | |
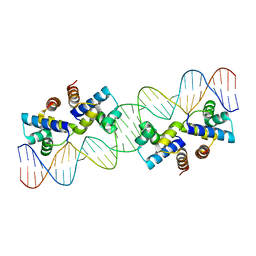
4X4H
 
 | |
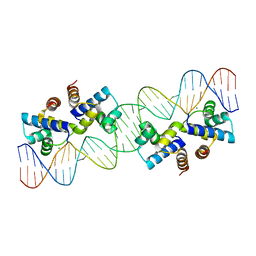
4X4B
 
 | |
4X4D
 
 | |
4X4C
 
 | |
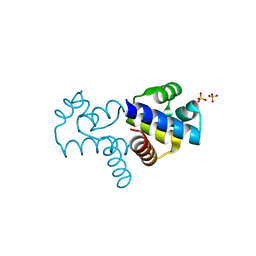
4FN3
 
 | |
4FBI
 
 | |
6HQK
 
 | | Crystal structure of GcoA F169A bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQR
 
 | | Crystal structure of GcoA F169H bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQM
 
 | | Crystal structure of GcoA F169I bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQS
 
 | | Crystal structure of GcoA F169S bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQP
 
 | | Crystal structure of GcoA F169V bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4XNN
 
 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Daphnia pulex | | Descriptor: | Cellobiohydrolase CHBI, GLYCEROL | | Authors: | Ebrahim, A, Hobdey, S.E, Podkaminer, K, Taylor II, L.E, Beckham, G.T, Decker, S.R, Himmel, M.E, Cragg, S.M, McGeehan, J.E. | | Deposit date: | 2015-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a GH7 Family Cellobiohydrolase from Daphnia pulex
To Be Published
|
|
5OGX
 
 | | Crystal structure of Amycolatopsis cytochrome P450 reductase GcoB. | | Descriptor: | BROMIDE ION, Cytochrome P450 reductase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-07-13 | | Release date: | 2018-07-04 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5OMU
 
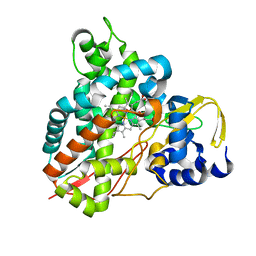 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5OMS
 
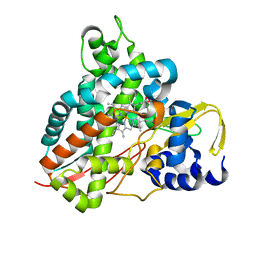 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with guaethol. | | Descriptor: | 2-ethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
6YCH
 
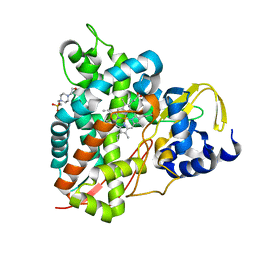 | | Crystal structure of GcoA T296A bound to guaiacol | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Ellis, E.S, DuBois, J.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCK
 
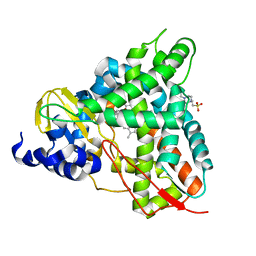 | | Crystal structure of GcoA T296A bound to p-vanillin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCP
 
 | | Crystal structure of GcoA F169V bound to o-vanillin | | Descriptor: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCM
 
 | | Crystal structure of GcoA T296S bound to p-vanillin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCN
 
 | | Crystal structure of GcoA F169A bound to o-vanillin | | Descriptor: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCI
 
 | | Crystal structure of GcoA T296G bound to guaiacol | | Descriptor: | Aromatic O-demethylase, cytochrome P450 subunit, Guaiacol, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
