1FG7
 
 | |
1FC5
 
 | |
1FG3
 
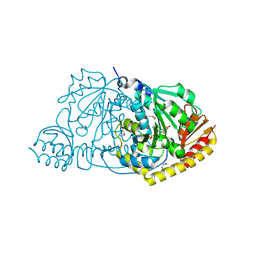 | | CRYSTAL STRUCTURE OF L-HISTIDINOL PHOSPHATE AMINOTRANSFERASE COMPLEXED WITH L-HISTIDINOL | | Descriptor: | HISTIDINOL PHOSPHATE AMINOTRANSFERASE, PHOSPHORIC ACID MONO-[2-AMINO-3-(3H-IMIDAZOL-4-YL)-PROPYL]ESTER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sivaraman, J, Cygler, M. | | Deposit date: | 2000-07-27 | | Release date: | 2001-08-22 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J.Mol.Biol., 311, 2001
|
|
2PQ4
 
 | |
3MPM
 
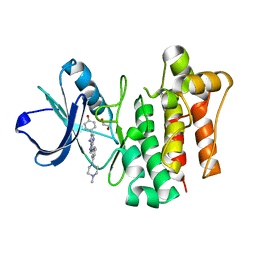 | | LCK complexed with a pyrazolopyrimidine | | Descriptor: | 1,2-ETHANEDIOL, 4-{(6R,7R)-7-amino-3-[3-(4-methylpiperazin-1-yl)phenyl]-6,7-dihydropyrazolo[1,5-a]pyrimidin-6-yl}phenol, Tyrosine-protein kinase Lck | | Authors: | Cowan-Jacob, S.W, Rummel, G. | | Deposit date: | 2010-04-27 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New pyrazolo[1,5a]pyrimidines as orally active inhibitors of Lck.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1NYT
 
 | | SHIKIMATE DEHYDROGENASE AroE COMPLEXED WITH NADP+ | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Roszak, A.W, Lapthorn, A.J. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of shikimate dehydrogenase AroE and its Paralog YdiB. A common structural framework for different activities.
J.Biol.Chem., 278, 2003
|
|
1O9B
 
 | | QUINATE/SHIKIMATE DEHYDROGENASE YDIB COMPLEXED WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB, PHOSPHATE ION | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2002-12-12 | | Release date: | 2003-02-01 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Shikimate Dehydrogenase Aroe and its Paralog Ydib: A Common Structural Framework for Different Activities
J.Biol.Chem., 278, 2003
|
|
1TVM
 
 | | NMR structure of enzyme GatB of the galactitol-specific phosphoenolpyruvate-dependent phosphotransferase system | | Descriptor: | PTS system, galactitol-specific IIB component | | Authors: | Volpon, L, Young, C.R, Lim, N.S, Iannuzzi, P, Cygler, M, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-06-29 | | Release date: | 2005-09-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the enzyme GatB of the galactitol-specific phosphoenolpyruvate-dependent phosphotransferase system and its interaction with GatA.
Protein Sci., 15, 2006
|
|
1U9T
 
 | |
1GZ0
 
 | | 23S RIBOSOMAL RNA G2251 2'O-METHYLTRANSFERASE RLMB | | Descriptor: | HYPOTHETICAL TRNA/RRNA METHYLTRANSFERASE YJFH | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2002-05-03 | | Release date: | 2002-10-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of the Rlmb 23S Rrna Methyltransferase Reveals a New Methyltransferase Fold with a Unique Knot
Structure, 10, 2002
|
|
1R8K
 
 | | PDXA PROTEIN; NAD-DEPENDENT DEHYDROGENASE/CARBOXYLASE; SUBUNIT OF PYRIDOXINE PHOSPHATE BIOSYNTHETIC PROTEIN PDXJ-PDXA [SALMONELLA TYPHIMURIUM] | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase 1, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Osipiuk, J, Quartey, P, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-27 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of NAD-dependent dehydrogenase/carboxylase of Salmonella typhimurium
to be published
|
|
1H1C
 
 | |
3UOY
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 1) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO, ... | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOV
 
 | | Crystal Structure of OTEMO (FAD bound form 1) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UP5
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 4) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UP4
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 3) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOZ
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 2) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOX
 
 | | Crystal Structure of OTEMO (FAD bound form 2) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
1P9K
 
 | | THE SOLUTION STRUCTURE OF YBCJ FROM E. COLI REVEALS A RECENTLY DISCOVERED ALFAL MOTIF INVOLVED IN RNA-BINDING | | Descriptor: | orf, hypothetical protein | | Authors: | Volpon, L, Lievre, C, Osborne, M.J, Gandhi, S, Iannuzzi, P, Larocque, R, Matte, A, Cygler, M, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-05-12 | | Release date: | 2003-11-25 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of YbcJ from Escherichia coli reveals a recently discovered alphaL motif involved in RNA binding.
J.Bacteriol., 185, 2003
|
|
1UU1
 
 | | Complex of Histidinol-phosphate aminotransferase (HisC) from Thermotoga maritima (Apo-form) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PHOSPHORIC ACID MONO-[2-AMINO-3-(3H-IMIDAZOL-4-YL)-PROPYL]ESTER | | Authors: | Vega, M.C, Fernandez, F.J, Lehman, F, Wilmanns, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Studies of the Catalytic Reaction Pathway of a Hyperthermophilic Histidinol-Phosphate Aminotransferase
J.Biol.Chem., 279, 2004
|
|
1UU0
 
 | | Histidinol-phosphate aminotransferase (HisC) from Thermotoga maritima (Apo-form) | | Descriptor: | HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Vega, M.C, Fernandez, F.J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Studies of the Catalytic Reaction Pathway of a Hyperthermophilic Histidinol-Phosphate Aminotransferase
J.Biol.Chem., 279, 2004
|
|
1UU2
 
 | | Histidinol-phosphate aminotransferase (HisC) from Thermotoga maritima (apo-form) | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, HISTIDINOL-PHOSPHATE AMINOTRANSFERASE | | Authors: | Vega, M.C, Fernandez, F.J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2003-12-13 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of the Catalytic Reaction Pathway of a Hyperthermophilic Histidinol-Phosphate Aminotransferase
J.Biol.Chem., 279, 2004
|
|
1KON
 
 | | CRYSTAL STRUCTURE OF E.COLI YEBC | | Descriptor: | Protein yebC | | Authors: | Jia, J, Smith, C, Lunin, V.V, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-12-21 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | UNPUBLISHED
TO BE PUBLISHED
|
|
