3W6G
 
 | | Structure of peroxiredoxin from anaerobic hyperthermophilic archaeon Pyrococcus horikoshii | | Descriptor: | CITRATE ANION, Probable peroxiredoxin | | Authors: | Nakamura, T, Mori, A, Niiyama, M, Matsumura, H, Tokuyama, C, Morita, J, Uegaki, K, Inoue, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of peroxiredoxin from the anaerobic hyperthermophilic archaeon Pyrococcus horikoshii
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3WQT
 
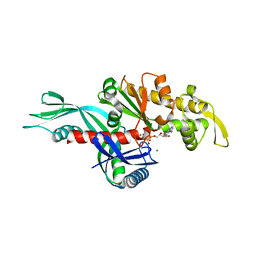 | | Staphylococcus aureus FtsA complexed with AMPPNP | | Descriptor: | CHLORIDE ION, Cell division protein FtsA, MAGNESIUM ION, ... | | Authors: | Fujita, J, Maeda, Y, Miyazaki, Y, Inoue, T, Matsumura, H. | | Deposit date: | 2014-02-01 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FtsA from Staphylococcus aureus
FEBS Lett., 588, 2014
|
|
3WQU
 
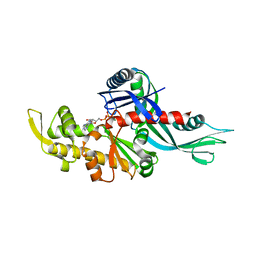 | | Staphylococcus aureus FtsA complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, MAGNESIUM ION | | Authors: | Fujita, J, Maeda, Y, Miyazaki, Y, Inoue, T, Matsumura, H. | | Deposit date: | 2014-02-01 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of FtsA from Staphylococcus aureus
FEBS Lett., 588, 2014
|
|
3AG6
 
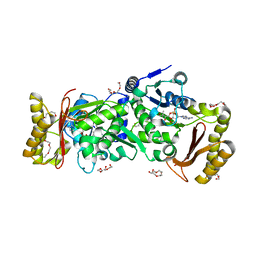 | | Crystal Structure of Pantothenate Synthetase from Staphylococcus aureus in complex with pantoyl adenylate | | Descriptor: | ACETIC ACID, PANTOYL ADENYLATE, Pantothenate synthetase, ... | | Authors: | Satoh, A, Konishi, S, Tamura, H, Stickland, H.G, Whitney, H.M, Smith, A.G, Matsumura, H, Inoue, T. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-induced closing of the active site revealed by the crystal structure of pantothenate synthetase from Staphylococcus aureus.
Biochemistry, 49, 2010
|
|
2ZXC
 
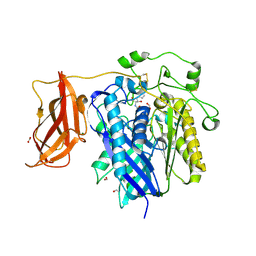 | | Ceramidase complexed with C2 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Okano, H, Inoue, T, Okino, N, Kakuta, Y, Matsumura, H, Ito, M. | | Deposit date: | 2008-12-22 | | Release date: | 2009-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase.
J.Biol.Chem., 284, 2009
|
|
3X2S
 
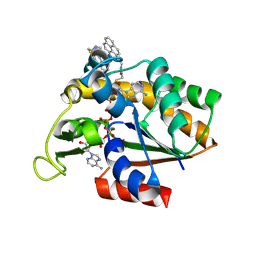 | | Crystal structure of pyrene-conjugated adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujii, A, Sekiguchi, Y, Matsumura, H, Inoue, T, Chung, W.-S, Hirota, S, Matsuo, T. | | Deposit date: | 2014-12-31 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Excimer Emission Properties on Pyrene-Labeled Protein Surface: Correlation between Emission Spectra, Ring Stacking Modes, and Flexibilities of Pyrene Probes.
Bioconjug.Chem., 26, 2015
|
|
3AGV
 
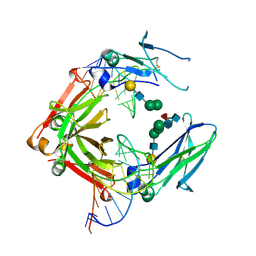 | | Crystal structure of a human IgG-aptamer complex | | Descriptor: | 5'-R(*GP*GP*AP*GP*GP*(UFT)P*GP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*GP*AP*AP*A*GP*GP*AP*AP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*A)-3', CALCIUM ION, Ig gamma-1 chain C region, ... | | Authors: | Nomura, Y, Sugiyama, S, Sakamoto, T, Miyakawa, S, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Nakamura, Y, Matsumura, H. | | Deposit date: | 2010-04-08 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational plasticity of RNA for target recognition as revealed by the 2.15 A crystal structure of a human IgG-aptamer complex
Nucleic Acids Res., 38, 2010
|
|
3AG5
 
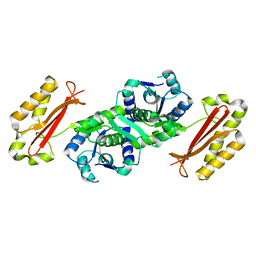 | | Crystal Structure of Pantothenate Synthetase from Staphylococcus aureus | | Descriptor: | Pantothenate synthetase | | Authors: | Satoh, A, Konishi, S, Tamura, H, Stickland, H.G, Whitney, H.M, Smith, A.G, Matsumura, H, Inoue, T. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate-induced closing of the active site revealed by the crystal structure of pantothenate synthetase from Staphylococcus aureus.
Biochemistry, 49, 2010
|
|
2YJN
 
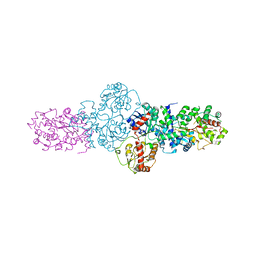 | | Structure of the glycosyltransferase EryCIII from the erythromycin biosynthetic pathway, in complex with its activating partner, EryCII | | Descriptor: | DTDP-4-KETO-6-DEOXY-HEXOSE 3,4-ISOMERASE, GLYCOSYLTRANSFERASE | | Authors: | Moncrieffe, M.C, Fernandez, M.J, Spiteller, D, Matsumura, H, Gay, N.J, Luisi, B.F, Leadlay, P.F. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Structure of the Glycosyltransferase Eryciii in Complex with its Activating P450 Homologue Erycii.
J.Mol.Biol., 415, 2012
|
|
2Z57
 
 | | Crystal structure of G56E-propeptide:S324A-subtilisin complex | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Pulido, M.A, Tanaka, S, Sringiew, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-06-29 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Requirement of left-handed glycine residue for high stability of the Tk-subtilisin propeptide as revealed by mutational and crystallographic analyses
J.Mol.Biol., 374, 2007
|
|
2Z56
 
 | | Crystal structure of G56S-propeptide:S324A-subtilisin complex | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Pulido, M.A, Tanaka, S, Sringiew, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-06-29 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Requirement of left-handed glycine residue for high stability of the Tk-subtilisin propeptide as revealed by mutational and crystallographic analyses
J.Mol.Biol., 374, 2007
|
|
2Z30
 
 | | Crystal structure of complex form between mat-Tk-subtilisin and Tk-propeptide | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Tanaka, S, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-29 | | Release date: | 2007-12-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Four new crystal structures of Tk-subtilisin in unautoprocessed, autoprocessed and mature forms: insight into structural changes during maturation
J.Mol.Biol., 372, 2007
|
|
2Z8Z
 
 | | Crystal structure of a platinum-bound S445C mutant of Pseudomonas sp. MIS38 lipase | | Descriptor: | CALCIUM ION, Lipase, PLATINUM (II) ION, ... | | Authors: | Angkawidjaja, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a family I.3 lipase from Pseudomonas sp. MIS38 in a closed conformation
FEBS Lett., 581, 2007
|
|
2Z58
 
 | | Crystal structure of G56W-propeptide:S324A-subtilisin complex | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Pulido, M.A, Tanaka, S, Sringiew, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-06-29 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Requirement of left-handed glycine residue for high stability of the Tk-subtilisin propeptide as revealed by mutational and crystallographic analyses
J.Mol.Biol., 374, 2007
|
|
2Z2X
 
 | | Crystal structure of mature form of Tk-subtilisin | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Tanaka, S, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-29 | | Release date: | 2007-12-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Four new crystal structures of Tk-subtilisin in unautoprocessed, autoprocessed and mature forms: insight into structural changes during maturation
J.Mol.Biol., 372, 2007
|
|
2Z2Z
 
 | | Crystal structure of unautoprocessed form of Tk-subtilisin soaked by 10mM CaCl2 | | Descriptor: | CALCIUM ION, Tk-subtilisin precursor | | Authors: | Tanaka, S, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-29 | | Release date: | 2007-12-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Four new crystal structures of Tk-subtilisin in unautoprocessed, autoprocessed and mature forms: insight into structural changes during maturation
J.Mol.Biol., 372, 2007
|
|
2ZCT
 
 | | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Hagihara, Y, Abe, M, Inoue, T, Yamamoto, T, Matsumura, H. | | Deposit date: | 2007-11-12 | | Release date: | 2008-05-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z8X
 
 | | Crystal structure of extracellular lipase from Pseudomonas sp. MIS38 | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Angkawidjaja, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-09-11 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of a family I.3 lipase from Pseudomonas sp. MIS38 in a closed conformation
FEBS Lett., 581, 2007
|
|
2Z2Y
 
 | | Crystal structure of autoprocessed form of Tk-subtilisin | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Tanaka, S, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-29 | | Release date: | 2007-12-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Four new crystal structures of Tk-subtilisin in unautoprocessed, autoprocessed and mature forms: insight into structural changes during maturation
J.Mol.Biol., 372, 2007
|
|
2ZRQ
 
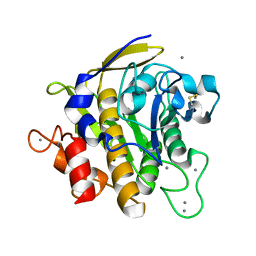 | | Crystal structure of S324A-subtilisin | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Tanaka, S, Takeuchi, Y, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2008-08-28 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Tk-subtilisin folded without propeptide: requirement of propeptide for acceleration of folding
Febs Lett., 582, 2008
|
|
2ZF5
 
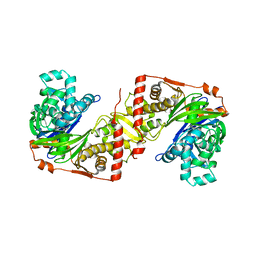 | | Crystal Structure of highly thermostable glycerol kinase from a hyperthermophilic archaeon | | Descriptor: | Glycerol kinase | | Authors: | Koga, Y, Katsumi, R, You, D.-J, Matsumura, H, Takano, K, Kanaya, S. | | Deposit date: | 2007-12-20 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of highly thermostable glycerol kinase from a hyperthermophilic archaeon in a dimeric form
Febs J., 275, 2008
|
|
2ZVD
 
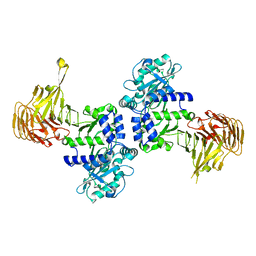 | | Crystal structure of Pseudomonas sp. MIS38 lipase in an open conformation | | Descriptor: | CALCIUM ION, Lipase | | Authors: | Angkawidjaja, C, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2008-11-05 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Crystallographic and MD Simulation Studies on the Mechanism of Interfacial Activation of a Family I.3 Lipase with Two Lids
J.Mol.Biol., 2010
|
|
3A3O
 
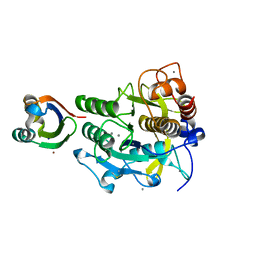 | | Crystal structure of complex between SA-subtilisin and Tk-propeptide with deletion of the five C-terminal residues | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Tanaka, S, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2009-06-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of the interactions critical for propeptide-catalyzed folding of Tk-subtilisin
J.Mol.Biol., 394, 2009
|
|
3A2W
 
 | | Peroxiredoxin (C50S) from Aeropytum pernix K1 (peroxide-bound form) | | Descriptor: | GLYCEROL, PEROXIDE ION, Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, F, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-06-04 | | Release date: | 2009-10-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3A70
 
 | | Crystal structure of Pseudomonas sp. MIS38 lipase in complex with diethyl phosphate | | Descriptor: | ACETATE ION, CALCIUM ION, DIETHYL PHOSPHONATE, ... | | Authors: | Angkawidjaja, C, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2009-09-10 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Crystallographic and MD Simulation Studies on the Mechanism of Interfacial Activation of a Family I.3 Lipase with Two Lids
J.Mol.Biol., 2010
|
|
