5JHC
 
 | |
4IHH
 
 | | Chasing Acyl Carrier Protein Through a Catalytic Cycle of Lipid A Production | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, ... | | Authors: | Masoudi, A, Raetz, C.R.H, Pemble, C.W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-11-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.132 Å) | | Cite: | Chasing acyl carrier protein through a catalytic cycle of lipid A production.
Nature, 505, 2014
|
|
4IHG
 
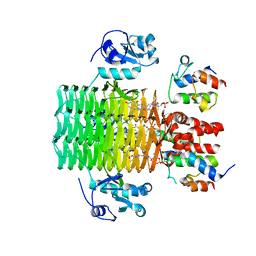 | | Chasing Acyl Carrier Protein Through a Catalytic Cycle of Lipid A Production | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, ... | | Authors: | Masoudi, A, Raetz, C.R.H, Pemble, C.W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-11-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Chasing acyl carrier protein through a catalytic cycle of lipid A production.
Nature, 505, 2014
|
|
4IHF
 
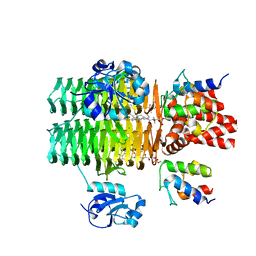 | | Chasing Acyl Carrier Protein Through a Catalytic Cycle of Lipid A Production | | Descriptor: | Acyl carrier protein, CHLORIDE ION, S-[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl] (3R)-3-hydroxytetradecanethioate, ... | | Authors: | Masoudi, A, Raetz, C.R.H, Pemble, C.W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-11-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chasing acyl carrier protein through a catalytic cycle of lipid A production.
Nature, 505, 2014
|
|
2AE2
 
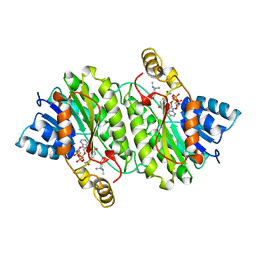 | | TROPINONE REDUCTASE-II COMPLEXED WITH NADP+ AND PSEUDOTROPINE | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (TROPINONE REDUCTASE-II), PSEUDOTROPINE | | Authors: | Yamashita, A, Kato, H, Wakatsuki, S, Tomizaki, T, Nakatsu, T, Nakajima, K, Hashimoto, T, Yamada, Y, Oda, J. | | Deposit date: | 1999-01-26 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of tropinone reductase-II complexed with NADP+ and pseudotropine at 1.9 A resolution: implication for stereospecific substrate binding and catalysis.
Biochemistry, 38, 1999
|
|
1IPE
 
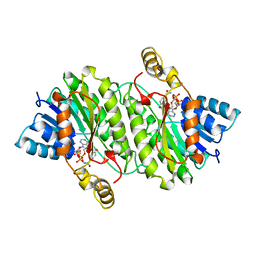 | | TROPINONE REDUCTASE-II COMPLEXED WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TROPINONE REDUCTASE-II | | Authors: | Yamashita, A, Endo, M, Higashi, T, Nakatsu, T, Yamada, Y, Oda, J, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-09 | | Release date: | 2003-06-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing Enzyme Structure Prior to Reaction Initiation: Tropinone Reductase-II-Substrate Complexes
BIOCHEMISTRY, 42, 2003
|
|
1IPF
 
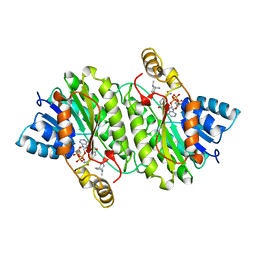 | | TROPINONE REDUCTASE-II COMPLEXED WITH NADPH AND TROPINONE | | Descriptor: | 8-METHYL-8-AZABICYCLO[3,2,1]OCTAN-3-ONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TROPINONE REDUCTASE-II | | Authors: | Yamashita, A, Endo, M, Higashi, T, Nakatsu, T, Yamada, Y, Oda, J, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-09 | | Release date: | 2003-06-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing Enzyme Structure Prior to Reaction Initiation: Tropinone Reductase-II-Substrate Complexes
BIOCHEMISTRY, 42, 2003
|
|
2KDY
 
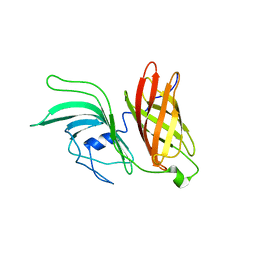 | | NMR structure of LP2086-B01 | | Descriptor: | Factor H binding protein variant B01_001 | | Authors: | Mascioni, A, Bentley, B.E, Camarda, R, Dilts, D.A, Fink, P, Gusarova, V, Hoiseth, S, Jacob, J, Lin, S.L, Malakian, K, McNeil, L.K, Mininni, T, Moy, F, Murphy, E, Novikova, E, Sigethy, S, Wen, Y, Zlotnick, G.W, Tsao, D.H.H. | | Deposit date: | 2009-01-20 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Immunogenic Properties of the Meningococcal Vaccine Candidate LP2086.
J.Biol.Chem., 284, 2009
|
|
2A65
 
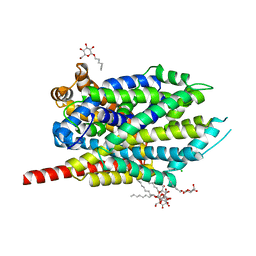 | | Crystal structure of LEUTAA, a bacterial homolog of Na+/Cl--dependent neurotransmitter transporters | | Descriptor: | CHLORIDE ION, LEUCINE, Na(+):neurotransmitter symporter (Snf family), ... | | Authors: | Yamashita, A, Singh, S.K, Kawate, T, Jin, Y, Gouaux, E. | | Deposit date: | 2005-07-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a bacterial homologue of Na(+)/Cl(-)-dependent neurotransmitter transporters.
Nature, 437, 2005
|
|
1KUW
 
 | | High-Resolution Structure and Localization of Amylin Nucleation Site in Detergent Micelles | | Descriptor: | Islet amyloid polypeptide | | Authors: | Mascioni, A, Porcelli, F, Ilangovan, U, Ramamoorthy, A, Veglia, G. | | Deposit date: | 2002-01-22 | | Release date: | 2003-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational preferences of the amylin nucleation site in SDS micelles: an NMR study.
Biopolymers, 69, 2003
|
|
1IZN
 
 | |
7VED
 
 | |
7BRQ
 
 | | Crystal structure of human FAM134B LIR fused to human GABARAP | | Descriptor: | GLYCEROL, Reticulophagy regulator 1,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Yamasaki, A, Noda, N.N. | | Deposit date: | 2020-03-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes.
Nat Commun, 11, 2020
|
|
7BRT
 
 | | Crystal structure of Sec62 LIR fused to GABARAP | | Descriptor: | Translocation protein SEC62,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Yamasaki, A, Noda, N.N. | | Deposit date: | 2020-03-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes.
Nat Commun, 11, 2020
|
|
7BRU
 
 | | Crystal structure of human RTN3 LIR fused to human GABARAP | | Descriptor: | PHOSPHATE ION, Reticulon-3,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Yamasaki, A, Noda, N.N. | | Deposit date: | 2020-03-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes.
Nat Commun, 11, 2020
|
|
7BRN
 
 | | Crystal structure of Atg40 AIM fused to Atg8 | | Descriptor: | 1,2-ETHANEDIOL, Autophagy-related protein 40,Autophagy-related protein 8, L-EPINEPHRINE | | Authors: | Yamasaki, A, Noda, N.N. | | Deposit date: | 2020-03-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes.
Nat Commun, 11, 2020
|
|
7YO8
 
 | |
7YO9
 
 | |
6AAG
 
 | |
6AAF
 
 | |
1JDM
 
 | | NMR Structure of Sarcolipin | | Descriptor: | Sarcolipin | | Authors: | Veglia, G, Mascioni, A. | | Deposit date: | 2001-06-14 | | Release date: | 2002-02-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and orientation of sarcolipin in lipid environments.
Biochemistry, 41, 2002
|
|
8B51
 
 | | Usutu virus methyltransferase domain in complex with sinefungin | | Descriptor: | GLYCEROL, GLYCINE, RNA-directed RNA polymerase NS5, ... | | Authors: | Ferrero, D.S, Albentosa Gonzalez, L, Mas, A, Verdaguer, N. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure and function of the NS5 methyltransferase domain from Usutu virus.
Antiviral Res., 208, 2022
|
|
8B52
 
 | | Usutu virus methyltransferase domain in complex with sinefungin | | Descriptor: | GLYCEROL, Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Ferrero, D.S, Albentosa Gonzalez, L, Mas, A, Verdaguer, N. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and function of the NS5 methyltransferase domain from Usutu virus.
Antiviral Res., 208, 2022
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8HPK
 
 | | Crystal structure of the bacterial oxalate transporter OxlT in an oxalate-bound occluded form | | Descriptor: | Fab fragment Heavy chein, Fab fragment Light chain, OXALATE ION, ... | | Authors: | Shimamura, T, Hirai, T, Yamashita, A. | | Deposit date: | 2022-12-12 | | Release date: | 2023-02-15 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanism of oxalate transporter OxlT in an oxalate-degrading bacterium in the gut microbiota.
Nat Commun, 14, 2023
|
|
