3HCC
 
 | | Crystal Structure of hPNMT in Complex With anti-9-amino-5-(trifluromethyl) benzonorbornene and AdoHcy | | Descriptor: | (1S,4R,9S)-5-(trifluoromethyl)-1,2,3,4-tetrahydro-1,4-methanonaphthalen-9-amine, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L, Gee, C.L, Puri, M. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
3BD2
 
 | |
3HCF
 
 | |
3HCE
 
 | | Crystal Structure of E185D hPNMT in Complex With Octopamine and AdoHcy | | Descriptor: | 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
3HCA
 
 | | Crystal Structure of E185Q hPNMT in Complex With Octopamine and AdoHcy | | Descriptor: | 1,2-ETHANEDIOL, 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, Phenylethanolamine N-methyltransferase, ... | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
3HCB
 
 | | Crystal Structure of hPNMT in Complex With Noradrenochrome and AdoHcy | | Descriptor: | (3S)-3-hydroxy-2,3-dihydro-1H-indole-5,6-dione, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L, Gee, C.L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
3HCD
 
 | | Crystal Structure of hPNMT in Complex With Noradrenaline and AdoHcy | | Descriptor: | L-NOREPINEPHRINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
4I1K
 
 | | Crystal Structure of VRN1 (Residues 208-341) | | Descriptor: | B3 domain-containing transcription factor VRN1, CHLORIDE ION | | Authors: | King, G, Chanson, A.H, McCallum, E.J, Ohme-Takagi, M, Byriel, K, Hill, J.M, Martin, J.L, Mylne, J.S. | | Deposit date: | 2012-11-21 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Arabidopsis B3 Domain Protein VERNALIZATION1 (VRN1) Is Involved in Processes Essential for Development, with Structural and Mutational Studies Revealing Its DNA-binding Surface.
J.Biol.Chem., 288, 2013
|
|
3UX3
 
 | | Crystal Structure of Domain-Swapped Fam96a minor dimer | | Descriptor: | ACETATE ION, MIP18 family protein FAM96A, ZINC ION | | Authors: | Chen, K.-E, Kobe, B, Martin, J.L. | | Deposit date: | 2011-12-03 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mammalian DUF59 protein Fam96a forms two distinct types of domain-swapped dimer.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UX2
 
 | |
4K6X
 
 | | Crystal structure of disulfide oxidoreductase from Mycobacterium tuberculosis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Disulfide oxidoreductase | | Authors: | Premkumar, L, Martin, J.L. | | Deposit date: | 2013-04-16 | | Release date: | 2013-10-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Rv2969c, essential for optimal growth in Mycobacterium tuberculosis, is a DsbA-like enzyme that interacts with VKOR-derived peptides and has atypical features of DsbA-like disulfide oxidases.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4MCU
 
 | |
4MLY
 
 | | Disulfide isomerase from multidrug resistance IncA/C related integrative and conjugative elements in oxidized state (P21 space group) | | Descriptor: | 1,3-BUTANEDIOL, DsbP | | Authors: | Premkumar, L, Kurth, F, Neyer, S, Martin, J.L. | | Deposit date: | 2013-09-06 | | Release date: | 2013-12-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | The Multidrug Resistance IncA/C Transferable Plasmid Encodes a Novel Domain-swapped Dimeric Protein-disulfide Isomerase.
J.Biol.Chem., 289, 2014
|
|
4ML6
 
 | |
4ML1
 
 | |
1A98
 
 | | XPRTASE FROM E. COLI COMPLEXED WITH GMP | | Descriptor: | XANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
1A0M
 
 | | 1.1 ANGSTROM CRYSTAL STRUCTURE OF A-CONOTOXIN [TYR15]-EPI | | Descriptor: | ALPHA-CONOTOXIN [TYR15]-EPI | | Authors: | Hu, S.-H, Loughnan, M, Miller, R, Weeks, C.M, Blessing, R.H, Alewood, P.F, Lewis, R.J, Martin, J.L. | | Deposit date: | 1997-12-03 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A resolution crystal structure of [Tyr15]EpI, a novel alpha-conotoxin from Conus episcopatus, solved by direct methods.
Biochemistry, 37, 1998
|
|
1A95
 
 | | XPRTASE FROM E. COLI COMPLEXED WITH MG:CPRPP AND GUANINE | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, BORIC ACID, GUANINE, ... | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
1A97
 
 | | XPRTASE FROM E. COLI COMPLEXED WITH GMP | | Descriptor: | BORIC ACID, GUANOSINE-5'-MONOPHOSPHATE, XANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
1AKG
 
 | | ALPHA-CONOTOXIN PNIB FROM CONUS PENNACEUS | | Descriptor: | ALPHA-CONOTOXIN PNIB | | Authors: | Hu, S.-H, Martin, J.L. | | Deposit date: | 1997-05-18 | | Release date: | 1998-05-20 | | Last modified: | 2015-06-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure at 1.1 A resolution of alpha-conotoxin PnIB: comparison with alpha-conotoxins PnIA and GI.
Biochemistry, 36, 1997
|
|
1A96
 
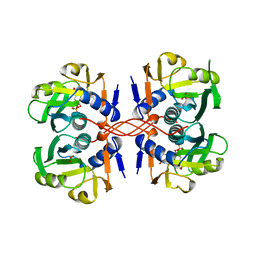 | | XPRTASE FROM E. COLI WITH BOUND CPRPP AND XANTHINE | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, BORIC ACID, MAGNESIUM ION, ... | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
1BED
 
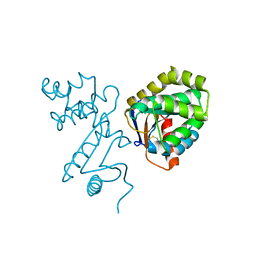 | | STRUCTURE OF DISULFIDE OXIDOREDUCTASE | | Descriptor: | DSBA OXIDOREDUCTASE | | Authors: | Hu, S.-H, Martin, J.L. | | Deposit date: | 1996-09-16 | | Release date: | 1997-10-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of TcpG, the DsbA protein folding catalyst from Vibrio cholerae.
J.Mol.Biol., 268, 1997
|
|
1CJM
 
 | | HUMAN SULT1A3 WITH SULFATE BOUND | | Descriptor: | PROTEIN (ARYL SULFOTRANSFERASE), SULFATE ION | | Authors: | Bidwell, L.M, Mcmanus, M.E, Gaedigk, A, Kakuta, Y, Negishi, M, Pedersen, L, Martin, J.L. | | Deposit date: | 1999-04-18 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human catecholamine sulfotransferase.
J.Mol.Biol., 293, 1999
|
|
1PEN
 
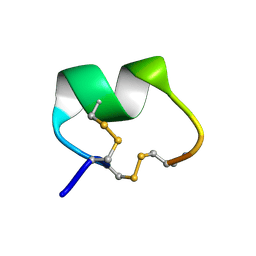 | | ALPHA-CONOTOXIN PNI1 | | Descriptor: | ALPHA-CONOTOXIN PNIA | | Authors: | Hu, S.-H, Gehrmann, J, Guddat, L.W, Alewood, P.F, Craik, D.J, Martin, J.L. | | Deposit date: | 1996-01-29 | | Release date: | 1997-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A crystal structure of the neuronal acetylcholine receptor antagonist, alpha-conotoxin PnIA from Conus pennaceus.
Structure, 4, 1996
|
|
1UTE
 
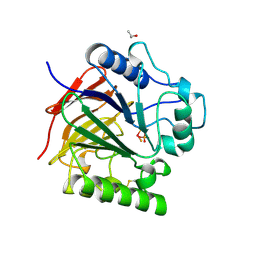 | | PIG PURPLE ACID PHOSPHATASE COMPLEXED WITH PHOSPHATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ISOPROPYL ALCOHOL, MU-OXO-DIIRON, ... | | Authors: | Guddat, L.W, Mcalpine, A, Hume, D, Hamilton, S, De Jersey, J, Martin, J.L. | | Deposit date: | 1999-01-18 | | Release date: | 1999-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of mammalian purple acid phosphatase.
Structure Fold.Des., 7, 1999
|
|
