3PUJ
 
 | | Crystal structure of the MUNC18-1 and SYNTAXIN4 N-Peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 1 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.313 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4DVC
 
 | | Structural and functional studies of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera toxin production | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Walden, P.M, Martin, J.L. | | Deposit date: | 2012-02-23 | | Release date: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A resolution crystal structure of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera-toxin production
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4GUX
 
 | | Crystal structure of trypsin:MCoTi-II complex | | Descriptor: | ACETATE ION, CALCIUM ION, Cationic trypsin, ... | | Authors: | King, G.J, Daly, N.L, Thorstholm, L, Greenwood, K.P, Rosengren, K.J, Heras, B, Craik, D.J, Martin, J.L. | | Deposit date: | 2012-08-30 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights into the role of the cyclic backbone in a squash trypsin inhibitor
J.Biol.Chem., 288, 2013
|
|
4OCF
 
 | |
4OCE
 
 | |
4OD7
 
 | |
4P3Y
 
 | |
4TKY
 
 | |
7RGV
 
 | |
2MBS
 
 | | NMR solution structure of oxidized KpDsbA | | Descriptor: | Thiol:disulfide interchange protein | | Authors: | Kurth, F, Rimmer, K, Premkumar, L, Mohanty, B, Duprez, W, Halili, M.A, Shouldice, S.R, Heras, B, Fairlie, D.P, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2013-08-03 | | Release date: | 2013-12-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comparative Sequence, Structure and Redox Analyses of Klebsiella pneumoniae DsbA Show That Anti-Virulence Target DsbA Enzymes Fall into Distinct Classes.
Plos One, 8, 2013
|
|
2OBF
 
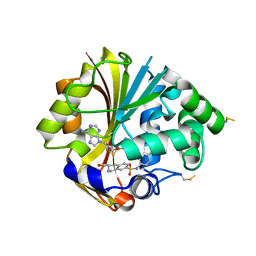 | | Structure of K57A hPNMT with inhibitor 3-Hydroxymethyl-7-(N-4-chlorophenylaminosulfonyl)-THIQ and AdoHcy (SAH) | | Descriptor: | (3R)-N-(4-CHLOROPHENYL)-3-(HYDROXYMETHYL)-1,2,3,4-TETRAHYDROISOQUINOLINE-7-SULFONAMIDE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2006-12-19 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme Adaptation to Inhibitor Binding: A Cryptic Binding Site in Phenylethanolamine N-Methyltransferase
J.Med.Chem., 50, 2007
|
|
2ONY
 
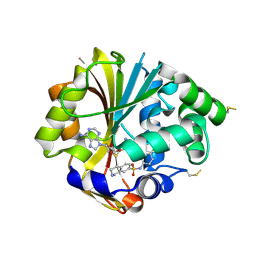 | | Structure of hPNMT with inhibitor 7-(N-4-chlorophenylaminosulfonyl)-THIQ and AdoHcy | | Descriptor: | N-(4-CHLOROPHENYL)-1,2,3,4-TETRAHYDROISOQUINOLINE-7-SULFONAMIDE, PHOSPHATE ION, Phenylethanolamine N-methyltransferase, ... | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2007-01-24 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enzyme Adaptation to Inhibitor Binding: A Cryptic Binding Site in Phenylethanolamine N-Methyltransferase
J.Med.Chem., 50, 2007
|
|
5TLQ
 
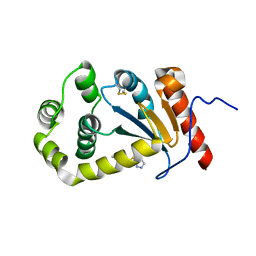 | | Model structure of the oxidized PaDsbA1 and 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine complex | | Descriptor: | 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, Thiol:disulfide interchange protein DsbA | | Authors: | Mohanty, B, Rimmer, K.A, McMahon, R.M, Headey, S.J, Vazirani, M, Shouldice, S.R, Coincon, M, Tay, S, Morton, C.J, Simpson, J.S, Martin, J.L, Scanlon, M.S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
5U3E
 
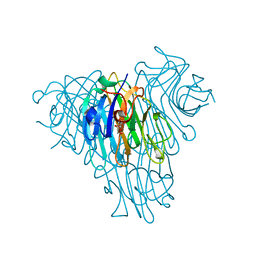 | | Crystal Structure of Native Lectin from Canavalia bonariensis Seeds (CaBo) complexed with alpha-methyl-D-mannoside | | Descriptor: | CALCIUM ION, Canavalia bonariensis seed lectin, MANGANESE (II) ION, ... | | Authors: | Silva, M.T.L, Osterne, V.J.S, Pinto-Junior, V.R, Santiago, M.Q, Araripe, D.A, Neco, A.H.B, Silva-Filho, J.C, Martins, J.L, Rocha, C.R.C, Leal, R.B, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Canavalia bonariensis lectin: Molecular bases of glycoconjugates interaction and antiglioma potential.
Int. J. Biol. Macromol., 106, 2018
|
|
5VYO
 
 | |
6BQX
 
 | | Crystal structure of Escherichia coli DsbA in complex with N-methyl-1-(4-phenoxyphenyl)methanamine | | Descriptor: | N-methyl-1-(4-phenoxyphenyl)methanamine, Thiol:disulfide interchange protein DsbA | | Authors: | Heras, B, Totsika, M, Paxman, J.J, Wang, G, Scanlon, M.J. | | Deposit date: | 2017-11-29 | | Release date: | 2017-12-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens.
Antioxid. Redox Signal., 29, 2018
|
|
9GPB
 
 | |
1BL0
 
 | | MULTIPLE ANTIBIOTIC RESISTANCE PROTEIN (MARA)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*TP*GP*CP*CP*AP*CP*GP*TP*TP*TP*TP*GP*CP*TP*AP*AP*AP*TP* CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*TP*AP*GP*CP*AP*AP*AP*AP*CP*GP*TP*GP*GP*CP*AP* TP*C)-3'), PROTEIN (MULTIPLE ANTIBIOTIC RESISTANCE PROTEIN) | | Authors: | Davies, S, Rhee, R.G, Martin, J.L, Rosner, D.R. | | Deposit date: | 1998-07-22 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6WI6
 
 | | Crystal structure of plantacyclin B21AG | | Descriptor: | MALONATE ION, Plantacyclin B21AG | | Authors: | Smith, A.T, Gor, M.C, Vezina, B, McMahon, R, King, G, Panjikar, S, Rehm, B, Martin, J. | | Deposit date: | 2020-04-08 | | Release date: | 2021-01-06 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and site-directed mutagenesis of circular bacteriocin plantacyclin B21AG reveals cationic and aromatic residues important for antimicrobial activity.
Sci Rep, 10, 2020
|
|
7JVE
 
 | | Crystal structure of Salmonella enterica Typhimurium BcfH | | Descriptor: | 1,2-ETHANEDIOL, DsbA family protein, MAGNESIUM ION, ... | | Authors: | Subedi, P, Heras, B, Hor, L, Paxman, J.J. | | Deposit date: | 2020-08-21 | | Release date: | 2021-04-21 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Salmonella enterica BcfH Is a Trimeric Thioredoxin-Like Bifunctional Enzyme with Both Thiol Oxidase and Disulfide Isomerase Activities.
Antioxid.Redox Signal., 35, 2021
|
|
4WET
 
 | | Crystal structure of E.Coli DsbA in complex with compound 16 | | Descriptor: | 1,2-ETHANEDIOL, N-({4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazol-5-yl}carbonyl)-L-tyrosine, SODIUM ION, ... | | Authors: | Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8EQQ
 
 | | Crystal structure of E.coli DsbA mutant E37A | | Descriptor: | CITRATE ANION, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
8EOC
 
 | | Crystal structure of E.coli DsbA mutant E24A/K58A | | Descriptor: | COPPER (II) ION, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-03 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
8EQP
 
 | | Crystal structure of E.coli DsbA mutant E24A/E37A/K58A | | Descriptor: | CITRATE ANION, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
8EQR
 
 | | Crystal structure of E.coli DsbA mutant E24A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
