7KNV
 
 | | Solution NMR structure of CDHR3 extracellular domain EC1 | | Descriptor: | CALCIUM ION, Cadherin-related family member 3 | | Authors: | Lee, W, Tonelli, M, Frederick, R.O, Watters, K.E, Markley, J.L, Palmenberg, A.C. | | Deposit date: | 2020-11-06 | | Release date: | 2021-02-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Determination of the CDHR3 Rhinovirus-C Binding Domain, EC1
Viruses, 13, 2021
|
|
7M98
 
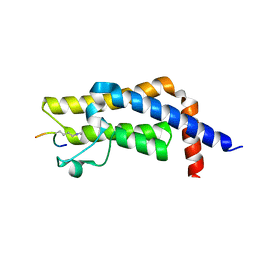 | | ATAD2 bromodomain complexed with histone H4K5ac (res 1-10) ligand | | Descriptor: | ATPase family AAA domain-containing protein 2, Histone H4 | | Authors: | Malone, K.L, Phillips, M, Nix, J.C, Glass, K.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Coordination of Di-Acetylated Histone Ligands by the ATAD2 Bromodomain.
Int J Mol Sci, 22, 2021
|
|
1FTG
 
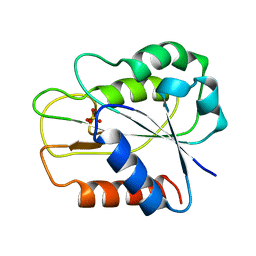 | |
2KYQ
 
 | | 1H, 15N, 13C chemical shifts and structure of CKR-brazzein | | Descriptor: | Defensin-like protein | | Authors: | Dittli, S.M, Assadi-Porter, F.M, Rao, H, Tonelli, M. | | Deposit date: | 2010-06-07 | | Release date: | 2011-05-18 | | Last modified: | 2011-11-30 | | Method: | SOLUTION NMR | | Cite: | Structural role of the terminal disulfide bond in the sweetness of brazzein.
Chem Senses, 36, 2011
|
|
2L07
 
 | | 1H, 13C, and 15N chemical shifts and structure of brazzein-derived peptide CKR-PNG | | Descriptor: | Defensin-like protein | | Authors: | Dittli, S.M, Assadi-Porter, F.M, Rao, H, Tonelli, M. | | Deposit date: | 2010-06-30 | | Release date: | 2011-06-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural role of the terminal disulfide bond in the sweetness of brazzein.
Chem Senses, 36, 2011
|
|
2LE4
 
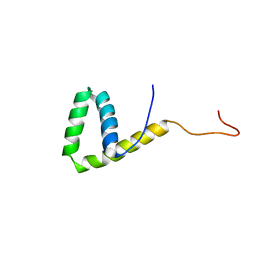 | | Solution structure of the HMG box DNA-binding domain of human stem cell transcription factor Sox2 | | Descriptor: | Transcription factor SOX-2 | | Authors: | Sahu, S.C, Markley, J.L, Tonelli, M, Bahrami, A, Eghbalnia, H.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2011-06-06 | | Release date: | 2011-07-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HMG box DNA-binding domain of human stem cell transcription factor Sox2
To be Published
|
|
2BF3
 
 | | Crystal structure of a toluene 4-monooxygenase catalytic effector protein variant missing ten N-terminal residues (delta-N10 T4moD) | | Descriptor: | HEPTANE-1,2,3-TRIOL, TOLUENE-4-MONOOXYGENASE SYSTEM PROTEIN D | | Authors: | Lountos, G.T, Mitchell, K.H, Studts, J.M, Fox, B.G, Orville, A.M. | | Deposit date: | 2004-12-03 | | Release date: | 2005-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures and Functional Studies of T4Mod, the Toluene 4-Monooxygenase Catalytic Effector Protein
Biochemistry, 44, 2005
|
|
2BF2
 
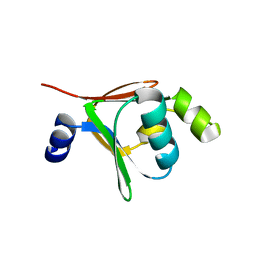 | | Crystal structure of native toluene-4-monooxygenase catalytic effector protein, T4moD | | Descriptor: | TOLUENE-4-MONOOXYGENASE SYSTEM PROTEIN D | | Authors: | Lountos, G.T, Mitchell, K.H, Studts, J.M, Fox, B.G, Orville, A.M. | | Deposit date: | 2004-12-03 | | Release date: | 2005-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures and Functional Studies of T4Mod, the Toluene 4-Monooxygenase Catalytic Effector Protein
Biochemistry, 44, 2005
|
|
2BF5
 
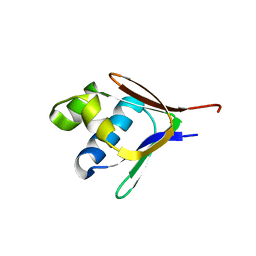 | | Crystal structure of a toluene 4-monooxygenase catalytic effector protein variant missing four N-terminal residues (delta-N4 T4moD) | | Descriptor: | TOLUENE-4-MONOOXYGENASE SYSTEM PROTEIN D | | Authors: | Lountos, G.T, Mitchell, K.H, Studts, J.M, Fox, B.G, Orville, A.M. | | Deposit date: | 2004-12-03 | | Release date: | 2005-05-19 | | Last modified: | 2016-12-28 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structures and Functional Studies of T4Mod, the Toluene 4-Monooxygenase Catalytic Effector Protein
Biochemistry, 44, 2005
|
|
2G0Q
 
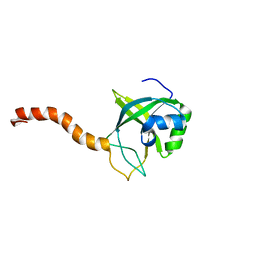 | | Solution structure of At5g39720.1 from Arabidopsis thaliana | | Descriptor: | AT5G39720.1 protein | | Authors: | Volkman, B.F, Peterson, F.C, Lytle, B.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-13 | | Release date: | 2006-02-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Arabidopsis thaliana protein At5g39720.1, a member of the AIG2-like protein family.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2GKD
 
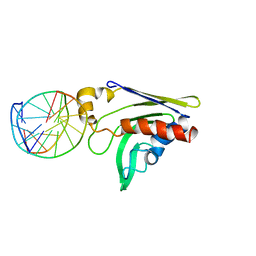 | |
2IL4
 
 | | Crystal structure of At1g77540-Coenzyme A Complex | | Descriptor: | COENZYME A, Protein At1g77540 | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
1PUT
 
 | | AN NMR-DERIVED MODEL FOR THE SOLUTION STRUCTURE OF OXIDIZED PUTIDAREDOXIN, A 2FE, 2-S FERREDOXIN FROM PSEUDOMONAS | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, PUTIDAREDOXIN | | Authors: | Pochapsky, T.C, Ye, X.M, Ratnaswamy, G, Lyons, T.A. | | Deposit date: | 1994-07-09 | | Release date: | 1994-09-30 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | An NMR-derived model for the solution structure of oxidized putidaredoxin, a 2-Fe, 2-S ferredoxin from Pseudomonas.
Biochemistry, 33, 1994
|
|
1PFD
 
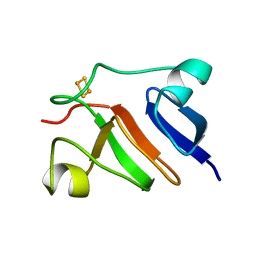 | | THE SOLUTION STRUCTURE OF HIGH PLANT PARSLEY [2FE-2S] FERREDOXIN, NMR, 18 STRUCTURES | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Im, S.-C, Liu, G, Luchinat, C, Sykes, A.G, Bertini, I. | | Deposit date: | 1998-05-05 | | Release date: | 1999-05-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of parsley [2Fe-2S]ferredoxin.
Eur.J.Biochem., 258, 1998
|
|
1QHE
 
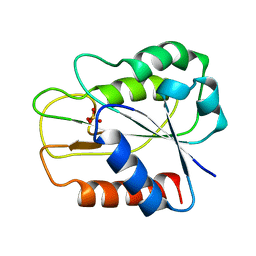 | |
1QO1
 
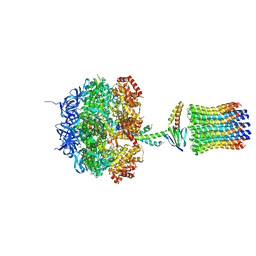 | | Molecular Architecture of the Rotary Motor in ATP Synthase from Yeast Mitochondria | | Descriptor: | ATP SYNTHASE ALPHA CHAIN, ATP SYNTHASE BETA CHAIN, ATP SYNTHASE DELTA CHAIN, ... | | Authors: | Stock, D, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1999-11-01 | | Release date: | 1999-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Molecular Architecture of the Rotary Motor in ATP Synthase
Science, 286, 1999
|
|
7W8H
 
 | |
7W8E
 
 | |
2MMI
 
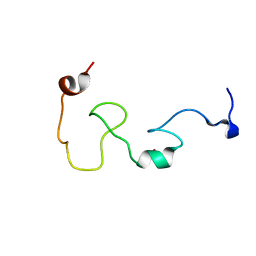 | | Mengovirus Leader: Structural Characterization of the Mengovirus Leader Protein Bound to Ran GTPase by Nuclear Magnetic Resonance | | Descriptor: | Leader protein, ZINC ION | | Authors: | Bacot-Davis, V.R, Cornilescu, C.C, Markley, J.L, Palmenberg, A.C. | | Deposit date: | 2014-03-15 | | Release date: | 2014-10-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2M7U
 
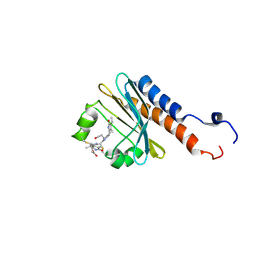 | | Blue Light-Absorbing State of TePixJ, an Active Cyanobacteriochrome Domain | | Descriptor: | Methyl-accepting chemotaxis protein, Phycoviolobilin, blue light-absorbing form | | Authors: | Cornilescu, G, Cornilescu, C.C, Burgie, S.E, Walker, J.M, Markley, J.L, Ulijasz, A.T, Vierstra, R.D. | | Deposit date: | 2013-05-01 | | Release date: | 2014-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Flexibility Variations Between the Ground and Photoactivated States of a Blue/Green Light-Absorbing Cyanobacteriochrome
To be Published
|
|
2M2B
 
 | | NMR structure of the RRM2 domain of the protein RBM10 from Homo sapiens | | Descriptor: | RNA-binding protein 10 | | Authors: | Serrano, P, Geralt, M, Dutta, S.K, Wuthrich, K, Wrobel, R.L, Makino, S, Misenhiemer, T.M, Markley, J.L, Fox, B.G, Joint Center for Structural Genomics (JCSG), Partnership for T-Cell Biology (TCELL), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2012-12-17 | | Release date: | 2013-01-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the RRM2 domain of the protein RBM10 from Homo sapiens
To be Published
|
|
2M7V
 
 | | Green Light-Absorbing State of TePixJ, an Active Cyanobacteriochrome Domain | | Descriptor: | Methyl-accepting chemotaxis protein, PHYCOVIOLOBILIN | | Authors: | Cornilescu, G, Cornilescu, C.C, Burgie, S.E, Walker, J.M, Markley, J.L, Ulijasz, A.T, Vierstra, R.D. | | Deposit date: | 2013-05-01 | | Release date: | 2014-01-01 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Structural and Flexibility Variations Between the Ground and Photoactivated States of a Blue/Green Light-Absorbing Cyanobacteriochrome
To be Published
|
|
2MMG
 
 | | Structural Characterization of the Mengovirus Leader Protein Bound to Ran GTPase by Nuclear Magnetic Resonance | | Descriptor: | GTP-binding nuclear protein Ran | | Authors: | Bacot-Davis, V.R, Palmenberg, A.C, Cornilescu, C.C, Markley, J.L. | | Deposit date: | 2014-03-15 | | Release date: | 2014-10-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2Q4A
 
 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At3g21360 | | Descriptor: | Clavaminate synthase-like protein At3g21360, FE (III) ION, SULFATE ION | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q44
 
 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At1g77540 | | Descriptor: | BROMIDE ION, Uncharacterized protein At1g77540 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
