8EUR
 
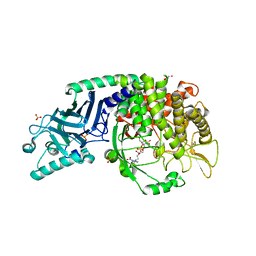 | | Co-crystal structure of Chaetomium glucosidase with compound 26 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{[2-nitro-4-(triazan-1-yl)phenyl]amino}ethyl (2-{[(1S,2S,3R,4S,5S)-2,3,4,5-tetrahydroxy-5-(hydroxymethyl)cyclohexyl]amino}ethyl)carbamate, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8ETO
 
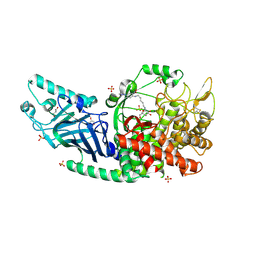 | | Co-crystal structure of Chaetomium glucosidase with compound 25 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-{[(5Z)-6-{[2-nitro-4-(2H-1,2,3-triazol-2-yl)phenyl]amino}hex-5-en-1-yl]amino}cyclohexane-1,2,3,4-tetrol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EPR
 
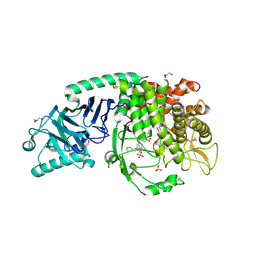 | | Co-crystal structure of Chaetomium glucosidase with compound 19 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[3-({[(5M)-3-(methanesulfonyl)-5-(pyridazin-3-yl)phenyl]amino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EPO
 
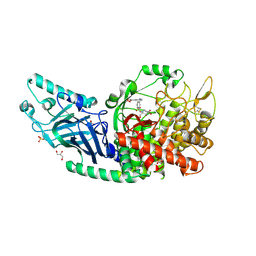 | | Co-crystal structure of Chaetomium glucosidase with compound 18 | | Descriptor: | (3P)-3-(5,6-dihydro-1,4-dioxin-2-yl)-5-{[(3-{[(2R,3R,4R,5S)-3,4,5-trihydroxy-2-(hydroxymethyl)piperidin-1-yl]methyl}phenyl)methyl]amino}benzonitrile, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EQ7
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 20 | | Descriptor: | (2R,3R,4R,5S)-1-[(3-{[3-bromo-5-(methanesulfonyl)anilino]methyl}phenyl)methyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8ETL
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 24 | | Descriptor: | (1S,2S,3R,4S,5S)-5-(butylamino)-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EUT
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 27 | | Descriptor: | (2R,3R,4R,5S)-1-[8-(furan-2-yl)octyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EUX
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 28 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{5-[4-(2-methoxyethyl)phenyl]pentyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EQX
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 21 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[6-({[(5M)-3-methyl-5-(1H-pyrrol-2-yl)phenyl]amino}methyl)pyridin-2-yl]methyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-10 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EPJ
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 17 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(3-{[4-(morpholin-4-yl)-2-nitroanilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8FWH
 
 | | Crystal structure of bivalent antibody Fab fragment of Anti-human LAG3 (22D2) | | Descriptor: | 1,2-ETHANEDIOL, Anti-human LAG3 (22D2) heavy chain, Anti-human LAG3 (22D2) light chain | | Authors: | Mishra, A.K, Agnihotri, P, Mariuzza, R.A. | | Deposit date: | 2023-01-22 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
1JWU
 
 | | Crystal Structure of the Complex of the MHC Class II Molecule HLA-DR1 (HA peptide 306-318) with the superantigen SEC3 Variant 3B2 | | Descriptor: | Enterotoxin type C-3, HA peptide, HLA class II histocompatibility antigen, ... | | Authors: | Sundberg, E.J, Andersen, P.S, Schlievert, P.M, Karjalainen, K, Mariuzza, R.A. | | Deposit date: | 2001-09-05 | | Release date: | 2003-07-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural, energetic, and functional analysis of a protein-protein interface at distinct stages of affinity
maturation
Structure, 11, 2003
|
|
1JWS
 
 | | Crystal Structure of the Complex of the MHC Class II Molecule HLA-DR1 (HA peptide 306-318) with the Superantigen SEC3 Variant 3B1 | | Descriptor: | Enterotoxin type C-3, HA peptide, HLA class II histocompatibility antigen, ... | | Authors: | Sundberg, E.J, Andersen, P.S, Schlievert, P.M, Karjalainen, K, Mariuzza, R.A. | | Deposit date: | 2001-09-05 | | Release date: | 2003-07-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural, energetic, and functional analysis of a protein-protein interface at distinct stages of affinity
maturation
Structure, 11, 2003
|
|
1JNE
 
 | | Crystal structure of imaginal disc growth factor-2 | | Descriptor: | Imaginal disc growth factor-2, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Varela, P.F, Llera, A.S, Mariuzza, R.A, Tormo, J. | | Deposit date: | 2001-07-23 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of imaginal disc growth factor-2. A member of a new family of growth-promoting glycoproteins from Drosophila melanogaster.
J.Biol.Chem., 277, 2002
|
|
1JWM
 
 | | Crystal Structure of the Complex of the MHC Class II Molecule HLA-DR1(HA peptide 306-318) with the Superantigen SEC3 | | Descriptor: | Enterotoxin type C-3, HA peptide, HLA class II histocompatibility antigen, ... | | Authors: | Sundberg, E.J, Andersen, P.S, Schlievert, P.M, Karjalainen, K, Mariuzza, R.A. | | Deposit date: | 2001-09-04 | | Release date: | 2003-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural, energetic, and functional analysis of a protein-protein interface at distinct stages of affinity
maturation
Structure, 11, 2003
|
|
1JND
 
 | | Crystal structure of imaginal disc growth factor-2 | | Descriptor: | Imaginal disc growth factor-2, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Varela, P.F, Llera, A.S, Mariuzza, R.A, Tormo, J. | | Deposit date: | 2001-07-23 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of imaginal disc growth factor-2. A member of a new family of growth-promoting glycoproteins from Drosophila melanogaster.
J.Biol.Chem., 277, 2002
|
|
1KLU
 
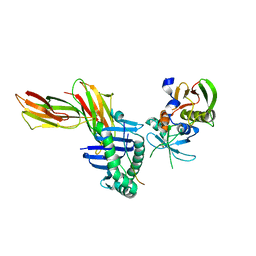 | | Crystal structure of HLA-DR1/TPI(23-37) complexed with staphylococcal enterotoxin C3 variant 3B2 (SEC3-3B2) | | Descriptor: | ENTEROTOXIN TYPE C-3, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, DR ALPHA CHAIN, ... | | Authors: | Sundberg, E.J, Sawicki, M.W, Andersen, P.S, Sidney, J, Sette, A, Mariuzza, R.A. | | Deposit date: | 2001-12-12 | | Release date: | 2002-08-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Minor structural changes in a mutated human melanoma antigen correspond to dramatically enhanced stimulation of a CD4+ tumor-infiltrating lymphocyte line.
J.Mol.Biol., 319, 2002
|
|
1KLG
 
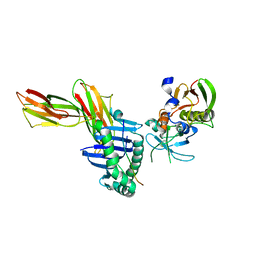 | | Crystal structure of HLA-DR1/TPI(23-37, Thr28-->Ile mutant) complexed with staphylococcal enterotoxin C3 variant 3B2 (SEC3-3B2) | | Descriptor: | ENTEROTOXIN TYPE C-3, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, DR ALPHA CHAIN, ... | | Authors: | Sundberg, E.J, Sawicki, M.W, Andersen, P.S, Sidney, J, Sette, A, Mariuzza, R.A. | | Deposit date: | 2001-12-11 | | Release date: | 2002-08-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Minor structural changes in a mutated human melanoma antigen correspond to dramatically enhanced stimulation of a CD4+ tumor-infiltrating lymphocyte line.
J.Mol.Biol., 319, 2002
|
|
1KTK
 
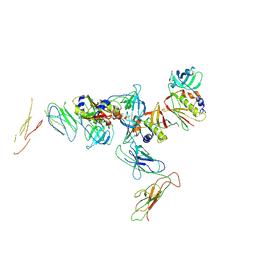 | | Complex of Streptococcal pyrogenic enterotoxin C (SpeC) with a human T cell receptor beta chain (Vbeta2.1) | | Descriptor: | Exotoxin type C, T-cell receptor beta chain | | Authors: | Sundberg, E.J, Li, H, Llera, A.S, McCormick, J.K, Tormo, J, Karjalainen, K, Schlievert, P.M, Mariuzza, R.A. | | Deposit date: | 2002-01-16 | | Release date: | 2002-06-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of two streptococcal superantigens bound to TCR beta chains reveal diversity in the architecture of T cell signaling complexes.
Structure, 10, 2002
|
|
1L0Y
 
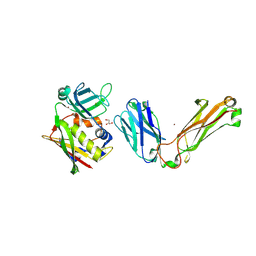 | | T cell receptor beta chain complexed with superantigen SpeA soaked with zinc | | Descriptor: | 14.3.d T cell receptor beta chain, Exotoxin type A, GLYCEROL, ... | | Authors: | Li, H, Sundberg, E.J, Mariuzza, R.A. | | Deposit date: | 2002-02-14 | | Release date: | 2002-04-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two streptococcal superantigens bound to TCR beta chains reveal diversity in the architecture of T cell signaling complexes.
Structure, 10, 2002
|
|
1XGP
 
 | |
1L0X
 
 | |
1XGU
 
 | | Structure for antibody HyHEL-63 Y33F mutant complexed with hen egg lysozyme | | Descriptor: | Lysozyme C, antibody kappa heavy chain, antibody kappa light chain | | Authors: | Li, Y, Huang, Y, Swaminathan, C.P, Smith-Gill, S.J, Mariuzza, R.A. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-06 | | Last modified: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Magnitude of the hydrophobic effect at central versus peripheral sites in protein-protein interfaces
Structure, 13, 2005
|
|
1XGR
 
 | | Structure for antibody HyHEL-63 Y33I mutant complexed with hen egg lysozyme | | Descriptor: | Lysozyme C, antibody kappa heavy chain, antibody kappa light chain | | Authors: | Li, Y, Huang, Y, Swaminathan, C.P, Smith-Gill, S.J, Mariuzza, R.A. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-06 | | Last modified: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Magnitude of the hydrophobic effect at central versus peripheral sites in protein-protein interfaces
Structure, 13, 2005
|
|
1XXG
 
 | |
