2Z27
 
 | | Thr109Ser dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
2Z2A
 
 | | Thr109Gly dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
2Z29
 
 | | Thr109Ala dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
1N51
 
 | | Aminopeptidase P in complex with the inhibitor apstatin | | Descriptor: | MANGANESE (II) ION, Xaa-Pro aminopeptidase, apstatin | | Authors: | Graham, S.C, Maher, M.J, Lee, M.H, Simmons, W.H, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-11-03 | | Release date: | 2003-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Escherichia coli aminopeptidase P in complex with the inhibitor apstatin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OV8
 
 | | Auracyanin B structure in space group, P65 | | Descriptor: | Auracyanin B, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Lee, M, Maher, M.J, Freeman, H.C, Guss, J.M. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Auracyanin B structure in space group P6(5).
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3GQB
 
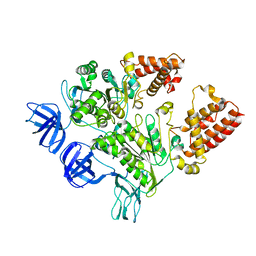 | | Crystal Structure of the A3B3 complex from V-ATPase | | Descriptor: | V-type ATP synthase alpha chain, V-type ATP synthase beta chain | | Authors: | Meher, M, Akimoto, S, Iwata, M, Nagata, K, Hori, Y, Yoshida, M, Yokoyama, S, Iwata, S, Yokoyama, K. | | Deposit date: | 2009-03-24 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of A(3)B(3) complex of V-ATPase from Thermus thermophilus.
Embo J., 28, 2009
|
|
4UTO
 
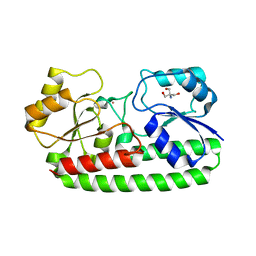 | | Crystal structure of pneumococcal surface antigen PsaA D280N in the Cd-bound, open state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
4UTP
 
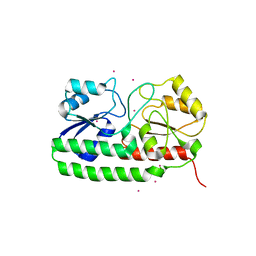 | | Crystal structure of pneumococcal surface antigen PsaA in the Cd- bound, closed state | | Descriptor: | CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
4Q5I
 
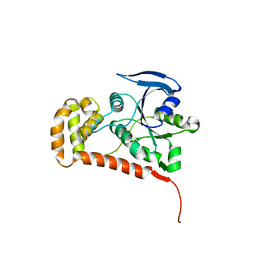 | | Crystal structure of a T151A mutant of the E. coli FeoB G-domain | | Descriptor: | Ferrous iron transport protein B, SULFATE ION | | Authors: | Jormakka, M, Guilfoyle, A.P, Deshpande, C.N, Maher, M. | | Deposit date: | 2014-04-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Exploring the correlation between the sequence composition of the nucleotide binding G5 loop of the FeoB GTPase domain (NFeoB) and intrinsic rate of GDP release.
Biosci.Rep., 34, 2014
|
|
6MWS
 
 | |
5K3X
 
 | | Crystal Structure of the sulfite dehydrogenase, SorT R78K mutant from Sinorhizobium meliloti | | Descriptor: | (MOLYBDOPTERIN-S,S)-OXO-MOLYBDENUM, GLYCEROL, Putative sulfite oxidase | | Authors: | Lee, M, McGrath, A, Maher, M. | | Deposit date: | 2016-05-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The central active site arginine in sulfite oxidizing enzymes alters kinetic properties by controlling electron transfer and redox interactions.
Biochim. Biophys. Acta, 1859, 2017
|
|
8CU8
 
 | |
2LEL
 
 | | Structure of Cu(I)Cu(II)-CopK from Cupriavidus metallidurans CH34 | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Copper resistance protein K | | Authors: | Hinds, M.G, Xiao, Z, Chong, L.X, Wedd, A.G. | | Deposit date: | 2011-06-16 | | Release date: | 2012-02-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of the cooperative binding of Cu(I) and Cu(II) to the CopK protein from Cupriavidus metallidurans CH34.
Biochemistry, 50, 2011
|
|
4NON
 
 | |
4Q00
 
 | |
1QHQ
 
 | | AURACYANIN, A BLUE COPPER PROTEIN FROM THE GREEN THERMOPHILIC PHOTOSYNTHETIC BACTERIUM CHLOROFLEXUS AURANTIACUS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (AURACYANIN), ... | | Authors: | Bond, C.S, Blankenship, R.E, Freeman, H.C, Guss, J.M, Maher, M, Selvaraj, F, Wilce, M.C.J, Willingham, K. | | Deposit date: | 1999-05-25 | | Release date: | 2001-03-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of auracyanin, a "blue" copper protein from the green thermophilic photosynthetic bacterium Chloroflexus aurantiacus.
J.Mol.Biol., 306, 2001
|
|
3W5I
 
 | |
3W5J
 
 | | Crystal structure of GDP-bound NfeoB from Gallionella capsiferriformans | | Descriptor: | Ferrous iron transport protein B, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Deshpande, C.N, McGrath, A.P, Jormakka, M. | | Deposit date: | 2013-01-30 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structure of an atypical FeoB G-domain reveals a putative domain-swapped dimer.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
