4L6G
 
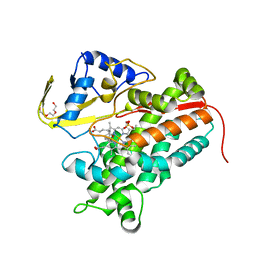 | | Crystal Structure of P450cin Y81F mutant, crystallized in 7 mM 1,8-cineole | | Descriptor: | 1,3,3-TRIMETHYL-2-OXABICYCLO[2.2.2]OCTANE, P450cin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Madrona, Y, Poulos, T.L. | | Deposit date: | 2013-06-12 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | P450cin active site water: implications for substrate binding and solvent accessibility.
Biochemistry, 52, 2013
|
|
4L77
 
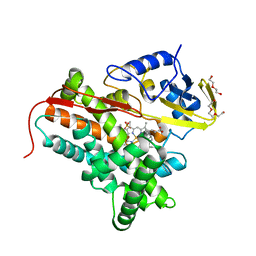 | | P450cin Active Site Water: Implications for Substrate Binding and Solvent Accessibility | | Descriptor: | 1,3,3-TRIMETHYL-2-OXABICYCLO[2.2.2]OCTANE, DI(HYDROXYETHYL)ETHER, P450cin, ... | | Authors: | Madrona, Y, Poulos, T.L. | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.379 Å) | | Cite: | P450cin active site water: implications for substrate binding and solvent accessibility.
Biochemistry, 52, 2013
|
|
4LHT
 
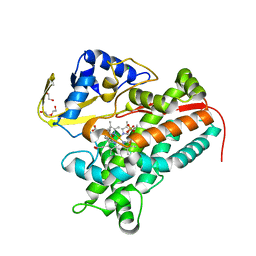 | | Crystal Structure of P450cin Y81F mutant, crystallized in 3 mM 1,8-cineole | | Descriptor: | 1,3,3-TRIMETHYL-2-OXABICYCLO[2.2.2]OCTANE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Madrona, Y, Poulos, T.L. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | P450cin active site water: implications for substrate binding and solvent accessibility.
Biochemistry, 52, 2013
|
|
4OXX
 
 | |
4FMX
 
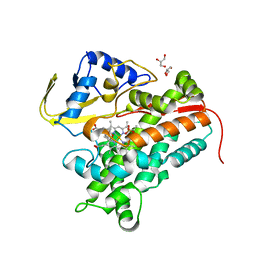 | | Crystal Structure of Substrate-Bound P450cin | | Descriptor: | 1,3,3-TRIMETHYL-2-OXABICYCLO[2.2.2]OCTANE, GLYCEROL, P450cin, ... | | Authors: | Madrona, Y, Tripathi, S.M, Huiying, L, Poulos, T.L. | | Deposit date: | 2012-06-18 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.554 Å) | | Cite: | Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation.
Biochemistry, 51, 2012
|
|
4FB2
 
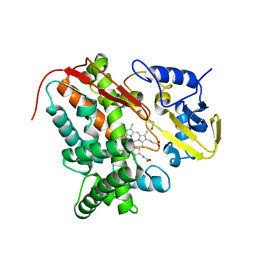 | | Crystal Structure of Substrate-Free P450cin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, P450cin, ... | | Authors: | Madrona, Y, Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2012-05-22 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation.
Biochemistry, 51, 2012
|
|
4FYZ
 
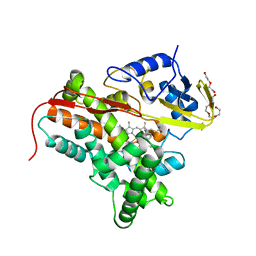 | | Crystal Structure of Nitrosyl Cytochrome P450cin | | Descriptor: | 1,3,3-TRIMETHYL-2-OXABICYCLO[2.2.2]OCTANE, DI(HYDROXYETHYL)ETHER, NITRIC OXIDE, ... | | Authors: | Madrona, Y, Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2012-07-05 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation.
Biochemistry, 51, 2012
|
|
4G3R
 
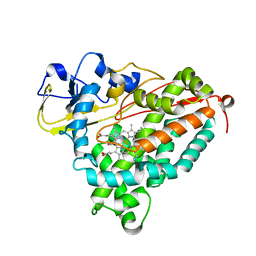 | | Crystal Structure of Nitrosyl Cytochrome P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, NITRIC OXIDE, ... | | Authors: | Madrona, Y, Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2012-07-15 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation.
Biochemistry, 51, 2012
|
|
4WPD
 
 | |
4YNR
 
 | | DosS GAFA Domain Reduced CO Bound Crystal Structure | | Descriptor: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, Redox sensor histidine kinase response regulator DevS | | Authors: | Madrona, Y. | | Deposit date: | 2015-03-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Analysis of cytochrome P450 CYP119 ligand-dependent conformational dynamics by two-dimensional NMR and X-ray crystallography.
J.Biol.Chem., 290, 2015
|
|
4YOF
 
 | | DosS GAFA Domain Reduced Nitric Oxide Bound Crystal Structure | | Descriptor: | NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE, Redox sensor histidine kinase response regulator DevS | | Authors: | Madrona, Y. | | Deposit date: | 2015-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of cytochrome P450 CYP119 ligand-dependent conformational dynamics by two-dimensional NMR and X-ray crystallography.
J.Biol.Chem., 290, 2015
|
|
4WQJ
 
 | |
4TT5
 
 | |
4TRI
 
 | |
4UAX
 
 | |
4TUV
 
 | |
