1ASP
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
1AVH
 
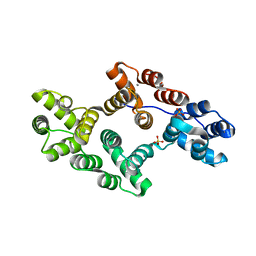 | | CRYSTAL AND MOLECULAR STRUCTURE OF HUMAN ANNEXIN V AFTER REFINEMENT. IMPLICATIONS FOR STRUCTURE, MEMBRANE BINDING AND ION CHANNEL FORMATION OF THE ANNEXIN FAMILY OF PROTEINS | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Huber, R, Berendes, R, Burger, A, Schneider, M, Karshikov, A, Luecke, H, Roemisch, J, Paques, E. | | Deposit date: | 1991-10-17 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal and molecular structure of human annexin V after refinement. Implications for structure, membrane binding and ion channel formation of the annexin family of proteins.
J.Mol.Biol., 223, 1992
|
|
1AVR
 
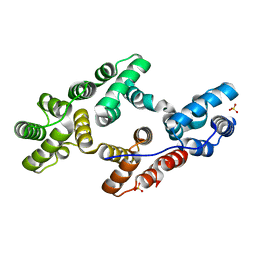 | | CRYSTAL AND MOLECULAR STRUCTURE OF HUMAN ANNEXIN V AFTER REFINEMENT. IMPLICATIONS FOR STRUCTURE, MEMBRANE BINDING AND ION CHANNEL FORMATION OF THE ANNEXIN FAMILY OF PROTEINS | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Huber, R, Berendes, R, Burger, A, Schneider, M, Karshikov, A, Luecke, H, Roemisch, J, Paques, E. | | Deposit date: | 1991-10-17 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal and molecular structure of human annexin V after refinement. Implications for structure, membrane binding and ion channel formation of the annexin family of proteins.
J.Mol.Biol., 223, 1992
|
|
1JV6
 
 | | BACTERIORHODOPSIN D85S/F219L DOUBLE MUTANT AT 2.00 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Rouhani, S, Cartailler, J.-P, Facciotti, M.T, Walian, P, Needleman, R, Lanyi, J.K, Glaeser, R.M, Luecke, H. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the D85S mutant of bacteriorhodopsin: model of an O-like photocycle intermediate.
J.Mol.Biol., 313, 2001
|
|
1JV7
 
 | | BACTERIORHODOPSIN O-LIKE INTERMEDIATE STATE OF THE D85S MUTANT AT 2.25 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Rouhani, S, Cartailler, J.-P, Facciotti, M.T, Walian, P, Needleman, R, Lanyi, J.K, Glaeser, R.M, Luecke, H. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the D85S mutant of bacteriorhodopsin: model of an O-like photocycle intermediate.
J.Mol.Biol., 313, 2001
|
|
1MEH
 
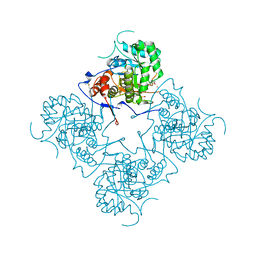 | |
1MEI
 
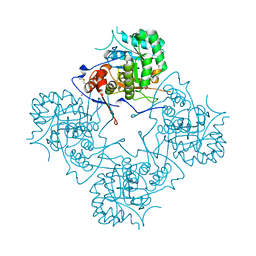 | |
1ME8
 
 | | Inosine Monophosphate Dehydrogenase (IMPDH) From Tritrichomonas Foetus with RVP bound | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, POTASSIUM ION, RIBAVIRIN MONOPHOSPHATE, ... | | Authors: | Prosise, G.L, Wu, J, Luecke, H. | | Deposit date: | 2002-08-08 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Tritrichomonas foetus Inosine Monophosphate Dehydrogenase
in Complex with the Inhibitor Ribavirin Monophosphate Reveals a
Catalysis-dependent Ion-binding Site
J.Biol.Chem., 277, 2002
|
|
1ME7
 
 | | Inosine Monophosphate Dehydrogenase (IMPDH) From Tritrichomonas Foetus with RVP and MOA bound | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, MYCOPHENOLIC ACID, POTASSIUM ION, ... | | Authors: | Prosise, G.L, Wu, J, Luecke, H. | | Deposit date: | 2002-08-08 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Tritrichomonas foetus Inosine Monophosphate Dehydrogenase
in Complex with the Inhibitor Ribavirin Monophosphate Reveals a Catalysis-dependent
Ion-binding Site
J.Biol.Chem., 277, 2002
|
|
1ME9
 
 | |
1MEW
 
 | |
2II2
 
 | | Crystal Structure of Alpha-11 Giardin | | Descriptor: | Alpha-11 giardin, SULFATE ION | | Authors: | Pathuri, P, Nguyen, E.T, Luecke, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Apo and Calcium-bound Crystal Structures of Alpha-11 Giardin, an Unusual Annexin from Giardia lamblia
J.Mol.Biol., 368, 2007
|
|
2II8
 
 | | Anabaena sensory rhodopsin transducer | | Descriptor: | Anabaena sensory rhodopsin transducer protein | | Authors: | Vogeley, L, Sineshchekov, O.A, Trivedi, V.D, Spudich, E.N, Spudich, J.L, Luecke, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the anabaena sensory rhodopsin transducer.
J.Mol.Biol., 367, 2007
|
|
2IIC
 
 | |
2IIA
 
 | | Anabaena sensory rhodopsin transducer | | Descriptor: | Sensory rhodopsin transducer protein | | Authors: | Vogeley, L, Sineshchekov, O.A, Trivedi, V.D, Spudich, E.N, Spudich, J.L, Luecke, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the anabaena sensory rhodopsin transducer.
J.Mol.Biol., 367, 2007
|
|
2OCJ
 
 | |
4HBC
 
 | |
4CSO
 
 | | The structure of OrfY from Thermoproteus tenax | | Descriptor: | ORFY PROTEIN, TRANSCRIPTION FACTOR | | Authors: | Zeth, K, Hagemann, A, Siebers, B, Martin, J, Lupas, A.N. | | Deposit date: | 2014-03-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
1IXI
 
 | |
5HZD
 
 | | RNA Editing TUTase 1 from Trypanosoma brucei | | Descriptor: | 3' terminal uridylyl transferase, CHLORIDE ION, SULFATE ION, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-02 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
5I49
 
 | | RNA Editing TUTase 1 from Trypanosoma brucei in complex with UTP analog UMPNPP | | Descriptor: | 3' terminal uridylyl transferase, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-11 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
5IDO
 
 | | RNA Editing TUTase 1 from Trypanosoma brucei in complex with UTP | | Descriptor: | 3' terminal uridylyl transferase, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Thore, S, Rajappa, L.T. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Nucleic Acids Res., 44, 2016
|
|
1Z17
 
 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein with bound ligand isoleucine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ISOLEUCINE, Leu/Ile/Val-binding protein | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
1Z18
 
 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein with bound valine | | Descriptor: | CADMIUM ION, Leu/Ile/Val-binding protein, VALINE | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
1Z16
 
 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein with bound leucine | | Descriptor: | CADMIUM ION, LEUCINE, Leu/Ile/Val-binding protein | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
