2HEX
 
 | |
6PA9
 
 | |
6PA5
 
 | |
6PA2
 
 | |
6PA4
 
 | |
1DOK
 
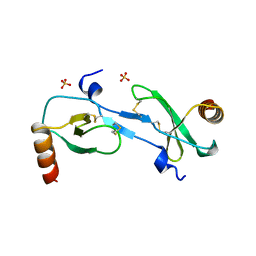 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, P-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1, SULFATE ION | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A, Domaille, P.J, Handel, T.M. | | Deposit date: | 1996-11-27 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
1DOL
 
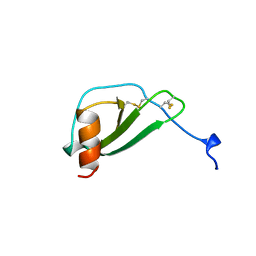 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, I-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1 | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A. | | Deposit date: | 1996-11-22 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
1G3P
 
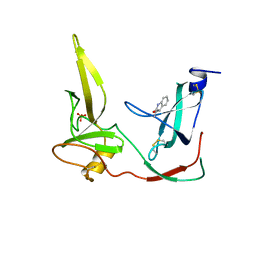 | | CRYSTAL STRUCTURE OF THE N-TERMINAL DOMAINS OF BACTERIOPHAGE MINOR COAT PROTEIN G3P | | Descriptor: | MINOR COAT PROTEIN, SULFATE ION | | Authors: | Lubkowski, J, Hennecke, F, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1997-12-22 | | Release date: | 1998-01-28 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p.
Nat.Struct.Biol., 5, 1998
|
|
2PM5
 
 | |
2PM1
 
 | |
5L04
 
 | |
8F1F
 
 | | Structure of K48-linked tri-ubiquitin in complex with cyclic peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Non-proteinogenic cyclic peptide (inhibitor), ... | | Authors: | Lubkowski, J, Fushman, D, Lemma, B. | | Deposit date: | 2022-11-05 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation.
Nat Commun, 14, 2023
|
|
5DUY
 
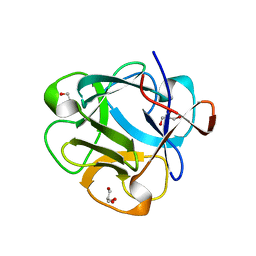 | | Structure of lectin from the sea mussel Crenomytilus grayanus | | Descriptor: | GLYCEROL, GalNAc/Gal-specific lectin | | Authors: | Lubkowski, J, Jakob, M, O'Keefe, B, Wlodawer, A. | | Deposit date: | 2015-09-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of a lectin from the sea mussel Crenomytilus grayanus (CGL).
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3HJ2
 
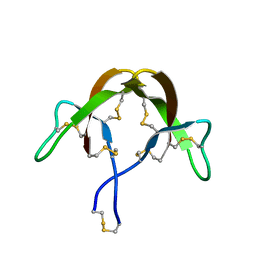 | |
3HJD
 
 | |
3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | Authors: | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8UOU
 
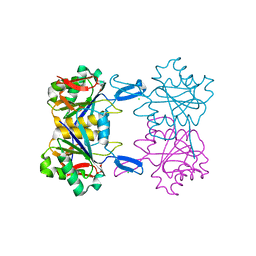 | | Structure of atypical asparaginase from Rhodospirillum rubrum in complex with L-Asp | | Descriptor: | ASPARTIC ACID, Asparaginase, CHLORIDE ION | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UPC
 
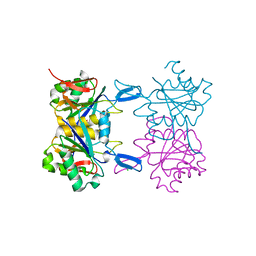 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K158M) | | Descriptor: | Asparaginase, CHLORIDE ION, GLYCEROL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP3
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F) | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, Asparaginase, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UOW
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21A) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Asparaginase | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP9
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K19Q) | | Descriptor: | 1,2-ETHANEDIOL, Asparaginase, CHLORIDE ION, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UOR
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K19E) | | Descriptor: | 1,2-ETHANEDIOL, Asparaginase, CHLORIDE ION | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP8
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F, complex with L-Asp) | | Descriptor: | ASPARTIC ACID, Asparaginase, CHLORIDE ION, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP7
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K19A) | | Descriptor: | Asparaginase, CHLORIDE ION | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UOO
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum | | Descriptor: | Asparaginase, CHLORIDE ION, GLYCEROL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
