6W36
 
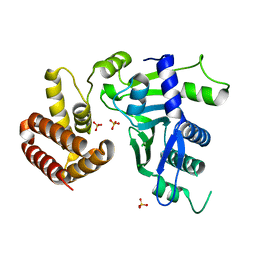 | | Crystal structure of FAM46C | | 分子名称: | SULFATE ION, Terminal nucleotidyltransferase 5C | | 著者 | Shang, G.J, Zhang, X.W, Chen, H, Lu, D.F. | | 登録日 | 2020-03-09 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.854 Å) | | 主引用文献 | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
6IZH
 
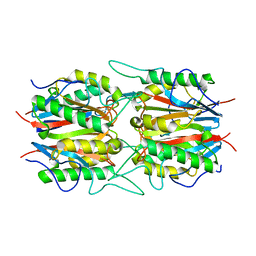 | |
5YPS
 
 | | The structural basis of histone chaperoneVps75 | | 分子名称: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | 登録日 | 2017-11-03 | | 公開日 | 2018-11-07 | | 最終更新日 | 2020-06-10 | | 実験手法 | X-RAY DIFFRACTION (2.097 Å) | | 主引用文献 | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
8IDN
 
 | | Cryo-EM structure of RBD/E77-Fab complex | | 分子名称: | E77 Fab heavy chain, E77 Fab light chain, Spike protein S1, ... | | 著者 | Lu, D.F, Zhang, Z.C. | | 登録日 | 2023-02-13 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | The structure of the RBD-E77 Fab complex reveals neutralization and immune escape of SARS-CoV-2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5ZB5
 
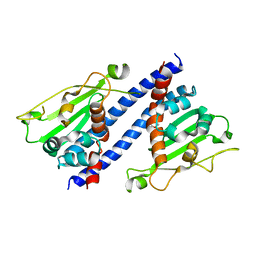 | | The structural basis of histone chaperoneVps75 | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NAP family histone chaperone vps75 | | 著者 | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | 登録日 | 2018-02-09 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.299 Å) | | 主引用文献 | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
6LJA
 
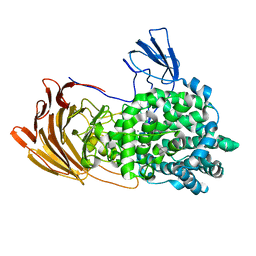 | | Crystal Structure of exoHep from Bacteroides intestinalis DSM 17393 complexed with disaccharide product | | 分子名称: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | 著者 | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | 登録日 | 2019-12-13 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.978 Å) | | 主引用文献 | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
6LJL
 
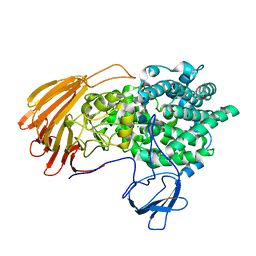 | | Crystal Structure of exoHep-Y390A/H555A complexed with a tetrasaccharide substrate | | 分子名称: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | 著者 | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | 登録日 | 2019-12-17 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
8IK0
 
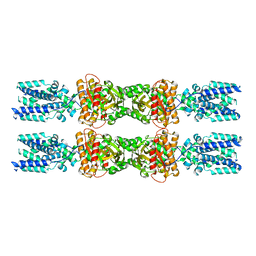 | |
8IK3
 
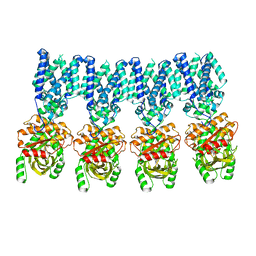 | |
7BYI
 
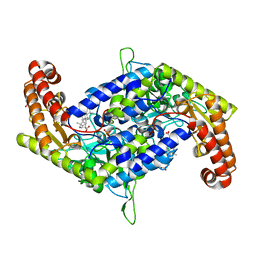 | | Structure of SHMT2 in complex with CBX | | 分子名称: | CARBENOXOLONE, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | 著者 | Li, L, Chen, Y, Lu, D, Zhang, C, Su, D. | | 登録日 | 2020-04-23 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Structure of SHMT2 with CBX
To Be Published
|
|
7DRV
 
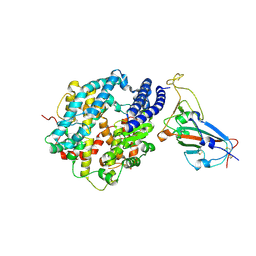 | | Structural basis of SARS-CoV-2-closely-related bat coronavirus RaTG13 to hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Liu, K.F, Pan, X.Q, Li, L.J, Feng, Y, Meng, Y.M, Zhang, Y.F, Wu, L.L, Chen, Q, Zheng, A.Q, Song, C.L, Jia, Y.F, Niu, S, Qiao, C.P, Zhao, X, Ma, D.L, Ma, X.P, Tan, S.G, Qi, J.X, Gao, G.F, Wang, Q.H. | | 登録日 | 2020-12-29 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species.
Cell, 184, 2021
|
|
8EQB
 
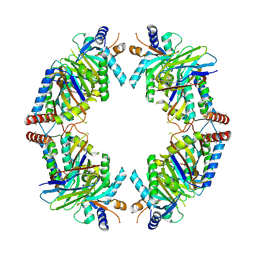 | | FAM46C/BCCIPalpha/Nanobody complex | | 分子名称: | Isoform 2 of BRCA2 and CDKN1A-interacting protein, Synthetic nanobody 1, Terminal nucleotidyltransferase 5C | | 著者 | Liu, S, Chen, H, Yin, Y, Bai, X, Zhang, X. | | 登録日 | 2022-10-07 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-04-26 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Inhibition of FAM46/TENT5 activity by BCCIP alpha adopting a unique fold.
Sci Adv, 9, 2023
|
|
8EXF
 
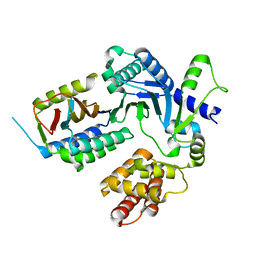 | |
8EXE
 
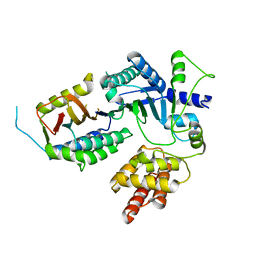 | |
6IYF
 
 | | Structure of pSTING complex | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-12-15 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.764 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6J2A
 
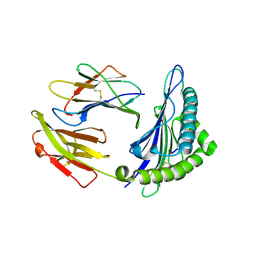 | | The structure of HLA-A*3003/NP44 | | 分子名称: | Beta-2-microglobulin, HLA-A*3003, NP44 | | 著者 | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, F.G, Lou, Y.L, Liu, W.J. | | 登録日 | 2018-12-31 | | 公開日 | 2019-09-25 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
6J1W
 
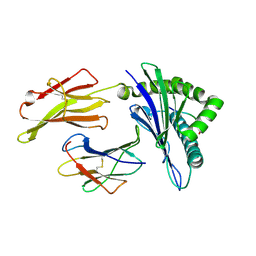 | | The structure of HLA-A*3001/RT313 | | 分子名称: | ALA-ILE-PHE-GLN-SER-SER-MET-THR-LYS, Beta-2-microglobulin, HLA-A*3001 | | 著者 | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, G.F, Lou, Y.L, Liu, W.J. | | 登録日 | 2018-12-29 | | 公開日 | 2019-09-25 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
6J29
 
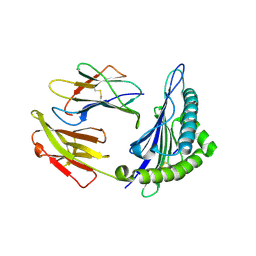 | | The structure of HLA-A*3003/MTB | | 分子名称: | Beta-2-microglobulin, HLA-A*3003, MTB | | 著者 | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, F.G, Lou, Y.L, Liu, W.J. | | 登録日 | 2018-12-31 | | 公開日 | 2019-09-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
6J1V
 
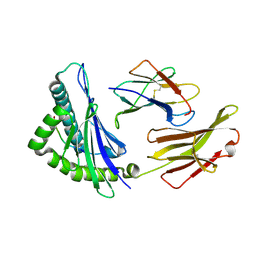 | | The structure of HLA-A*3003/RT313 | | 分子名称: | Beta-2-microglobulin, HLA-A*3003, RT313 | | 著者 | Zhu, S.Y, Liu, K.F, Chai, Y, Ding, C.M, Lv, J.X, Gao, G.F, Lou, Y.L, Liu, W.J. | | 登録日 | 2018-12-29 | | 公開日 | 2019-09-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Divergent Peptide Presentations of HLA-A*30 Alleles Revealed by Structures With Pathogen Peptides.
Front Immunol, 10, 2019
|
|
5XM2
 
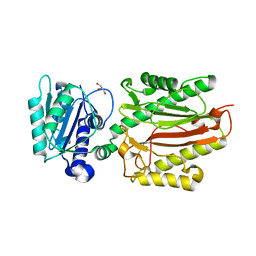 | | Human N-terminal domain of FACT complex subunit SPT16 | | 分子名称: | DI(HYDROXYETHYL)ETHER, FACT complex subunit SPT16, GLYCEROL | | 著者 | Xu, S, Li, H, Dou, Y, Chen, Y, Jiang, H, Lu, D, Wang, M, Su, D. | | 登録日 | 2017-05-12 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.187 Å) | | 主引用文献 | The structural basis of human Spt16 N-terminal domain interaction with histone (H3-H4)2tetramer.
Biochem.Biophys.Res.Commun., 508, 2019
|
|
5Y36
 
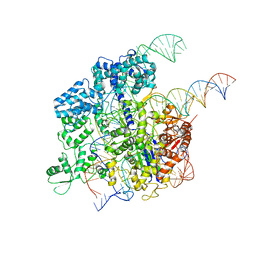 | | Cryo-EM structure of SpCas9-sgRNA-DNA ternary complex | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, complementary DNA strand, ... | | 著者 | Huang, Q, Li, G, Huai, C. | | 登録日 | 2017-07-27 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | Structural insights into DNA cleavage activation of CRISPR-Cas9 system
Nat Commun, 8, 2017
|
|
6A06
 
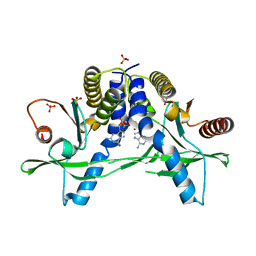 | | Structure of pSTING complex | | 分子名称: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A04
 
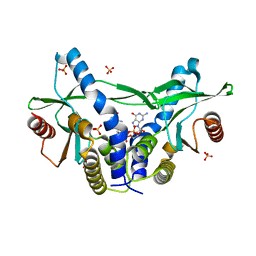 | | Structure of pSTING complex | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A05
 
 | | Structure of pSTING complex | | 分子名称: | 2-amino-9-[(2R,3R,3aR,5S,7aS,9R,10R,10aR,12R,14aS)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A03
 
 | | Structure of pSTING complex | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | 著者 | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | 登録日 | 2018-06-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.597 Å) | | 主引用文献 | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
