7C0G
 
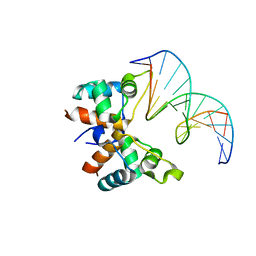 | |
3SQZ
 
 | |
3SR7
 
 | |
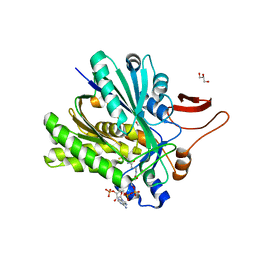
8HD5
 
 | |
9FR2
 
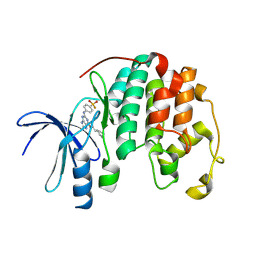 | |
3LEH
 
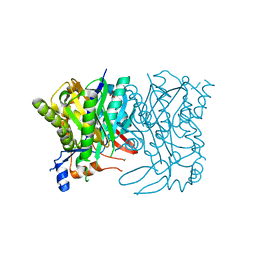 | |
4IXA
 
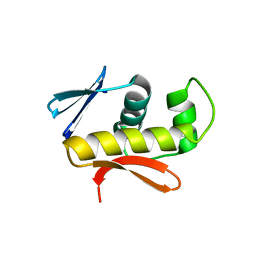 | | Structure of DNA-binding domain of the response regulator SaeR from Staphylococcus epidermidis | | Descriptor: | Response regulator SaeR | | Authors: | Chen, Y.R, Chen, S.C, Yang, C.S, Kuan, S.M, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-01-24 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of DNA-binding domain of the response regulator SaeR from Staphylococcus epidermidis
To be Published
|
|
4FKZ
 
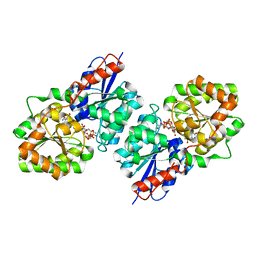 | | Crystal structure of Bacillus subtilis UDP-GlcNAc 2-epimerase in complex with UDP-GlcNAc and UDP | | Descriptor: | UDP-N-acetylglucosamine 2-epimerase, URIDINE-5'-DIPHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Yang, C.S, Chen, S.C, Kuan, S.M, Chen, Y.R, Liu, Y.H, Chen, Y. | | Deposit date: | 2012-06-14 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of Bacillus subtilis UDP-GlcNAc 2-epimerase in complex with UDP-GlcNAc and UDP
To be Published
|
|
4JIS
 
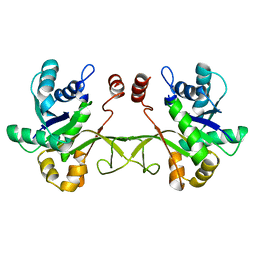 | | Crystal structure of ribitol 5-phosphate cytidylyltransferase (TarI) from Bacillus subtilis | | Descriptor: | ribitol-5-phosphate cytidylyltransferase | | Authors: | Yang, C.S, Chen, S.C, Chen, Y.R, Kuan, S.M, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-03-06 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Crystal structure of ribitol 5-phosphate cytidylyltransferase (TarI) from Bacillus subtilis
To be Published
|
|
5GNX
 
 | | The E171Q mutant structure of Bgl6 | | Descriptor: | Beta-glucosidase, GLYCEROL, PROPANOIC ACID, ... | | Authors: | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | Deposit date: | 2016-07-25 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
5GNZ
 
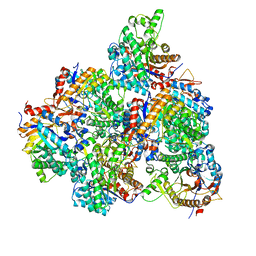 | | The M3 mutant structure of Bgl6 | | Descriptor: | Beta-glucosidase, GLYCEROL, beta-D-glucopyranose | | Authors: | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | Deposit date: | 2016-07-25 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
5GNY
 
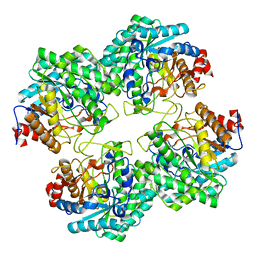 | | The structure of WT Bgl6 | | Descriptor: | Beta-glucosidase, beta-D-glucopyranose | | Authors: | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | Deposit date: | 2016-07-25 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
7VJQ
 
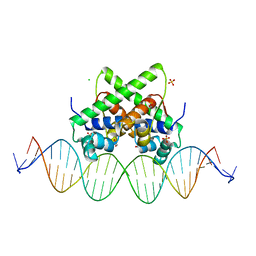 | | Pectobacterium phage ZF40 apo-aca2 complexed with 26bp DNA substrate | | Descriptor: | CHLORIDE ION, DNA (27-MER), GLYCEROL, ... | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7VJO
 
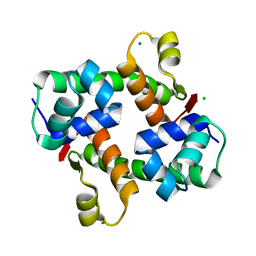 | | Pectobacterium phage ZF40 apo-Aca2 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, anti-CRISPR-associated protein Aca2 | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7VJN
 
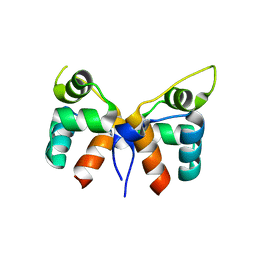 | |
7VJM
 
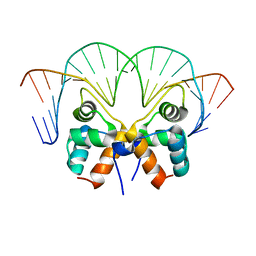 | | Aca1 in complex with 19bp palindromic DNA substrate | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*CP*AP*CP*AP*TP*TP*GP*TP*GP*CP*CP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*GP*GP*CP*AP*CP*AP*AP*TP*GP*TP*GP*CP*CP*TP*AP*A)-3'), anti-CRISPR-associated protein Aca1 | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7VJP
 
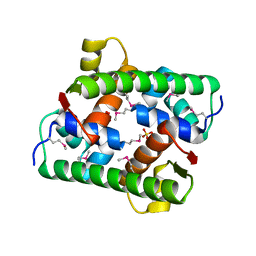 | | Selenomethionine-derived Pectobacterium phage ZF40 apo-Aca2 | | Descriptor: | SULFATE ION, anti-CRISPR-associated protein Aca2 | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7XTO
 
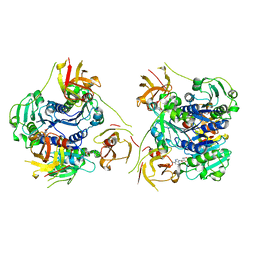 | |
7YF1
 
 | |
4ZRS
 
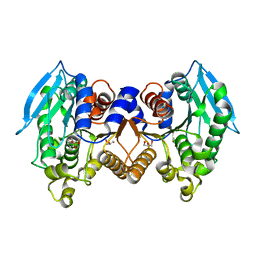 | | Crystal structure of a cloned feruloyl esterase from a soil metagenomic library | | Descriptor: | Esterase, GLYCEROL | | Authors: | Xie, W, Chen, R, Cao, L, Liu, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enhancing the Thermostability of Feruloyl Esterase EstF27 by Directed Evolution and the Underlying Structural Basis
J.Agric.Food Chem., 63, 2015
|
|
2FV5
 
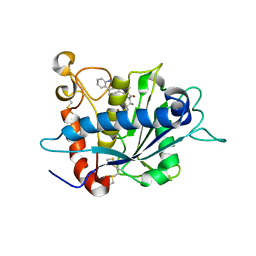 | | Crystal structure of TACE in complex with IK682 | | Descriptor: | (2R)-N-HYDROXY-2-[(3S)-3-METHYL-3-{4-[(2-METHYLQUINOLIN-4-YL)METHOXY]PHENYL}-2-OXOPYRROLIDIN-1-YL]PROPANAMIDE, ADAM 17, ZINC ION | | Authors: | Orth, P, Niu, X. | | Deposit date: | 2006-01-30 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IK682, a tight binding inhibitor of TACE.
Arch.Biochem.Biophys., 451, 2006
|
|
5FCC
 
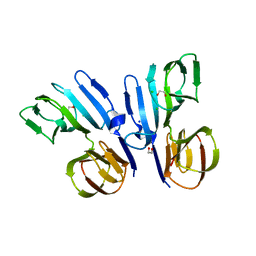 | | Structure of HutD from Pseudomonas fluorescens SBW25 (NaCl condition) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, HutD, ... | | Authors: | Johnston, J.M, Gerth, M.L, Baker, E.N, Lott, J.S, Rainey, P.B. | | Deposit date: | 2015-12-15 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of HutD from Pseudomonas fluorescens
To Be Published
|
|
4IWG
 
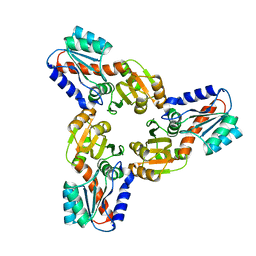 | | Crystal Structure of the Conserved Hypothetical Protein MJ0927 from Methanocaldococcus jannaschii (in C2221 form) | | Descriptor: | UPF0135 protein MJ0927 | | Authors: | Kuan, S.M, Chen, S.C, Yang, C.S, Chen, Y.R, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-01-23 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Crystal structure of a conserved hypothetical protein MJ0927 from Methanocaldococcus jannaschii reveals a novel quaternary assembly in the Nif3 family.
Biomed Res Int, 2014, 2014
|
|
4IWM
 
 | | Crystal Structure of the Conserved Hypothetical Protein MJ0927 from Methanocaldococcus jannaschii (in P21 form) | | Descriptor: | UPF0135 protein MJ0927 | | Authors: | Kuan, S.M, Chen, S.C, Yang, C.S, Chen, Y.R, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-01-24 | | Release date: | 2014-01-29 | | Last modified: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a conserved hypothetical protein MJ0927 from Methanocaldococcus jannaschii reveals a novel quaternary assembly in the Nif3 family.
Biomed Res Int, 2014, 2014
|
|
