7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (6.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YG7
 
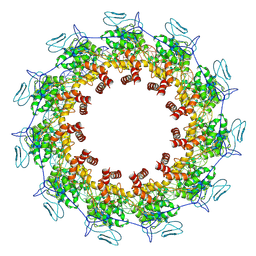 | | Structure of the Spring Viraemia of Carp Virus ribonucleoprotein Complex | | Descriptor: | Nucleoprotein, RNA (99-mer) | | Authors: | Liu, B, Wang, Z.X, Yang, T, Yu, D.Q, Ouyang, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Spring Viraemia of Carp Virus Ribonucleoprotein Complex Reveals Its Assembly Mechanism and Application in Antiviral Drug Screening.
J.Virol., 97, 2023
|
|
5YFE
 
 | |
6FI7
 
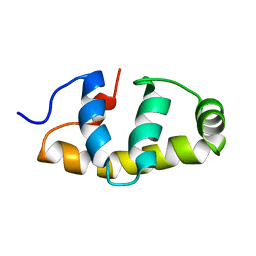 | | E.coli Sigma factor S (RpoS) Region 4 | | Descriptor: | RNA polymerase sigma factor RpoS | | Authors: | Liu, B, Matthews, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | T7 phage factor required for managing RpoS inEscherichia coli.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3UIY
 
 | |
3UIZ
 
 | |
6J4C
 
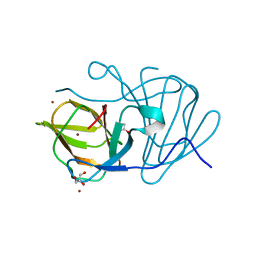 | | Crystal structure of MarH, an epimerase for biosynthesis of Maremycins in Streptomyces, under 10 mM ZnSO4 | | Descriptor: | ACETIC ACID, Cupin superfamily protein, GLYCEROL, ... | | Authors: | Hou, Y, Liu, B, Hu, K, Zhang, R. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis of the mechanism of beta-methyl epimerization by enzyme MarH.
Org.Biomol.Chem., 17, 2019
|
|
6J4D
 
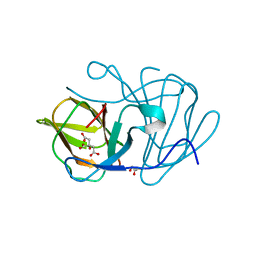 | | Crystal structure of MarH, an epimerase for biosynthesis of Maremycins in Streptomyces, under pH 4.7, without Zn | | Descriptor: | CITRATE ANION, Cupin superfamily protein, GLYCEROL | | Authors: | Hou, Y, Liu, B, Hu, K, Zhang, R. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for the Isomerization Mechanism of MarH
To Be Published
|
|
6J4B
 
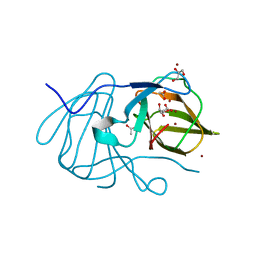 | | Crystal structure of MarH, an epimerase for biosynthesis of Maremycins in Streptomyces, under 400 mM Zinc acetate | | Descriptor: | ACETIC ACID, Cupin superfamily protein, GLYCEROL, ... | | Authors: | Hou, Y, Liu, B, Hu, K, Zhang, R. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis of the mechanism of beta-methyl epimerization by enzyme MarH.
Org.Biomol.Chem., 17, 2019
|
|
2ND4
 
 | |
2M6O
 
 | | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase | | Descriptor: | Uncharacterized protein | | Authors: | Liu, B, Tabib-Salazar, A, Doughty, P, Lewis, R, Ghosh, S, Parsy, M, Simpson, P, Matthews, S, Paget, M. | | Deposit date: | 2013-04-06 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase.
Nucleic Acids Res., 41, 2013
|
|
2M6P
 
 | |
6L6V
 
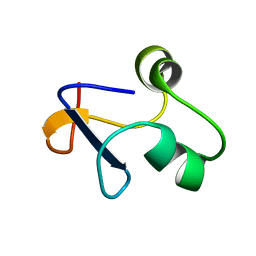 | | SPO1 Gp44 N-terminal region (1-55) | | Descriptor: | E3 protein | | Authors: | Liu, B, Wang, Z. | | Deposit date: | 2019-10-29 | | Release date: | 2021-05-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Bacteriophage DNA Mimic Protein Employs a Non-specific Strategy to Inhibit the Bacterial RNA Polymerase.
Front Microbiol, 12, 2021
|
|
7BY7
 
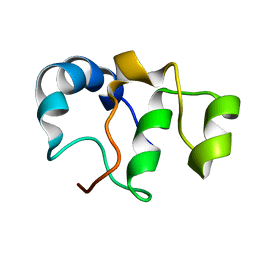 | | Bacteriophage SPO1 protein Gp46 | | Descriptor: | Putative gene 46 protein | | Authors: | Liu, B, Zhang, P. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Bacteriophage protein Gp46 is a cross-species inhibitor of nucleoid-associated HU proteins
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1APL
 
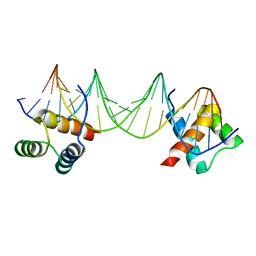 | | CRYSTAL STRUCTURE OF A MAT-ALPHA2 HOMEODOMAIN-OPERATOR COMPLEX SUGGESTS A GENERAL MODEL FOR HOMEODOMAIN-DNA INTERACTIONS | | Descriptor: | DNA (5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*CP*AP*TP*TP*TP*AP*C P*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*GP*TP*AP*AP*AP*TP*GP*AP*AP*TP*TP*A P*CP*AP*TP*G)-3'), PROTEIN (MAT-ALPHA2 HOMEODOMAIN) | | Authors: | Wolberger, C, Vershon, A.K, Liu, B, Johnson, A.D, Pabo, C.O. | | Deposit date: | 1993-10-04 | | Release date: | 1993-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions.
Cell(Cambridge,Mass.), 67, 1991
|
|
6WL5
 
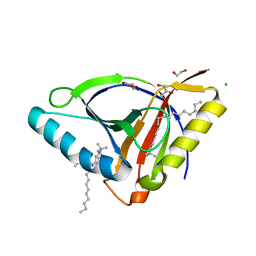 | | Crystal structure of EcmrR C-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, CETYL-TRIMETHYL-AMMONIUM, CHLORIDE ION, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-04-18 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLK
 
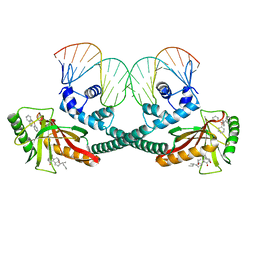 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-4nt | | Descriptor: | CHAPSO, MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLA
 
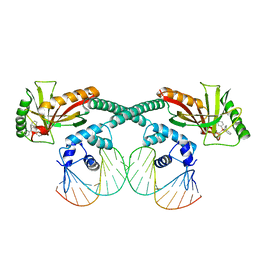 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-3nt | | Descriptor: | MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, synthetic non-template strand DNA (54-MER), ... | | Authors: | Yang, Y, Liu, C, Shi, W, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XL6
 
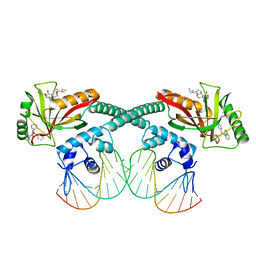 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPo | | Descriptor: | CHAPSO, MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XL9
 
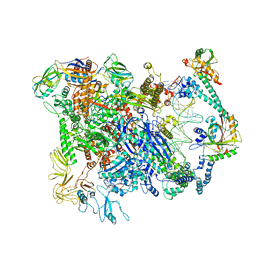 | | Cryo-EM structure of EcmrR-RNAP-promoter initial transcribing complex with 3-nt RNA transcript (EcmrR-RPitc-3nt) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Yang, Y, Liu, C, Shi, W, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLN
 
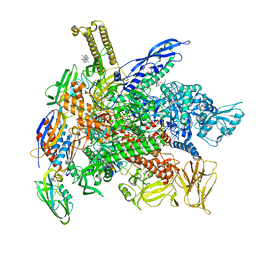 | | Cryo-EM structure of E. coli RNAP-DNA elongation complex 2 (RDe2) in EcmrR-dependent transcription | | Descriptor: | 9-nt RNA transcript, CHAPSO, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLJ
 
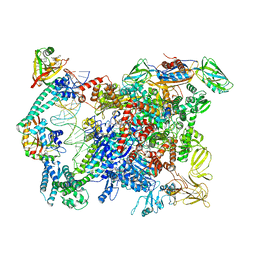 | | Cryo-EM structure of EcmrR-RNAP-promoter initial transcribing complex with 4-nt RNA transcript (EcmrR-RPitc-4nt) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLM
 
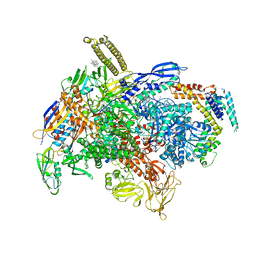 | | Cryo-EM structure of E.coli RNAP-DNA elongation complex 1 (RDe1) in EcmrR-dependent transcription | | Descriptor: | 9-nt RNA transcript, CHAPSO, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XL5
 
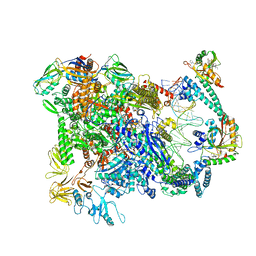 | | Cryo-EM structure of EcmrR-RNAP-promoter open complex (EcmrR-RPo) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLL
 
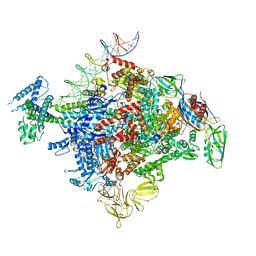 | | Cryo-EM structure of E. coli RNAP-promoter initial transcribing complex with 5-nt RNA transcript (RPitc-5nt) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
