6WTB
 
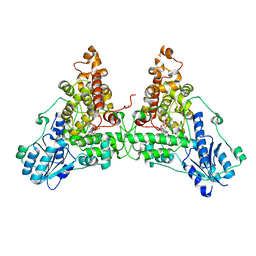 | | Sort-Tagged Drosophila Cryptochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Schneps, C.M, Crane, B.R. | | Deposit date: | 2020-05-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Tuning flavin environment to detect and control light-induced conformational switching in Drosophila cryptochrome.
Commun Biol, 4, 2021
|
|
8AR2
 
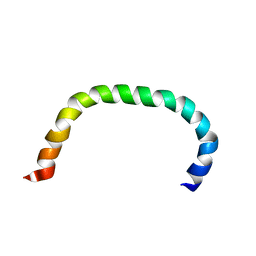 | | Solution structure of TLR5 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 5 | | Authors: | Shabalkina, A.V, Kornilov, F.D, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR3
 
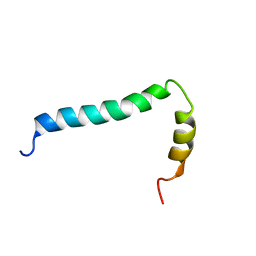 | | Solution structure of TLR9 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 9 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR1
 
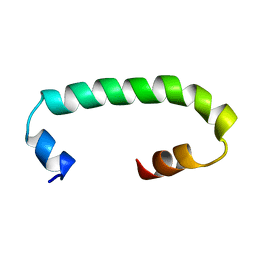 | | Solution structure of TLR3 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 3 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR0
 
 | | Solution structure of TLR2 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 2 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
5W0C
 
 | | Cytochrome P450 (CYP) 2C9 TCA007 Inhibitor Complex | | Descriptor: | Cytochrome P450 2C9, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Johnson, E.F, Hsu, M.-H. | | Deposit date: | 2017-05-30 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Determinants of the Inhibition of DprE1 and CYP2C9 by Antitubercular Thiophenes.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7NT7
 
 | |
7NUX
 
 | | Crystal structure of the TIR domain of human TLR1 (crystallised without ZN2+ ions) | | Descriptor: | Toll-like receptor 1 | | Authors: | Vakhrameev, D.D, Luginina, A.P, Shevtsov, M.B, Lushpa, V.A, Mineev, K.S, Borshchevskiy, V.I. | | Deposit date: | 2021-03-15 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Modulation of Toll-like receptor 1 intracellular domain structure and activity by Zn 2+ ions.
Commun Biol, 4, 2021
|
|
7NUW
 
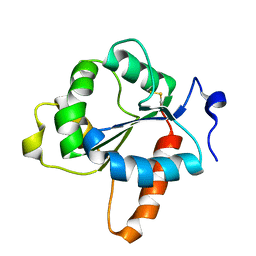 | | Crystal structure of the TIR domain of human TLR1 (crystallised with Zn2+ ions) | | Descriptor: | Toll-like receptor 1 | | Authors: | Vakhrameev, D.D, Luginina, A.P, Shevtsov, M.B, Lushpa, V.A, Mineev, K.S, Borshchevskiy, V.I. | | Deposit date: | 2021-03-15 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of Toll-like receptor 1 intracellular domain structure and activity by Zn 2+ ions.
Commun Biol, 4, 2021
|
|
4GL2
 
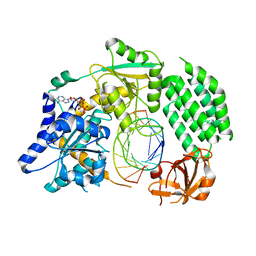 | | Structural Basis for dsRNA duplex backbone recognition by MDA5 | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(*AP*UP*CP*CP*GP*CP*GP*GP*CP*CP*CP*U)-3'), ... | | Authors: | Wu, B, Hur, S. | | Deposit date: | 2012-08-13 | | Release date: | 2013-01-09 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (3.557 Å) | | Cite: | Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5.
Cell(Cambridge,Mass.), 152, 2013
|
|
2WFK
 
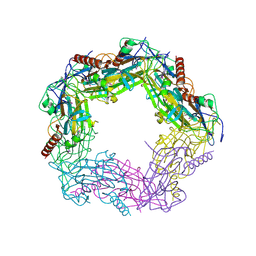 | | Calcium bound LipL32 | | Descriptor: | CALCIUM ION, LIPL32 | | Authors: | Tung, J.-Y, Yang, C.-W, Sun, Y.-J. | | Deposit date: | 2009-04-07 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Calcium Binds to Lipl32, a Lipoprotein from Pathogenic Leptospira, and Modulates Fibronectin Binding.
J.Biol.Chem., 285, 2010
|
|
7EI3
 
 | |
7EI4
 
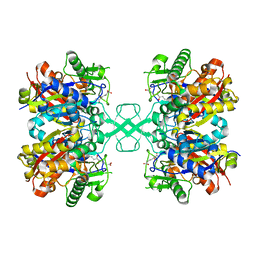 | | Crystal structure of MasL in complex with a novel covalent inhibitor, collimonin C | | Descriptor: | (6S,7R,9E)-6,7-bis(oxidanyl)hexadeca-9,15-dien-11,13-diynoic acid, Acetyl-CoA C-acyltransferase | | Authors: | Lin, C.C, Huang, K.F, Yang, Y.L. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Integrated omics approach to unveil antifungal bacterial polyynes as acetyl-CoA acetyltransferase inhibitors.
Commun Biol, 5, 2022
|
|
7FEA
 
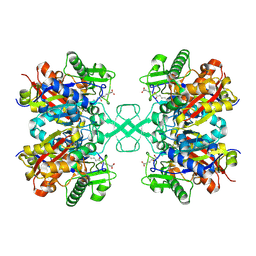 | | PY14 in complex with Col-D | | Descriptor: | (6~{R},7~{R},9~{E})-6,7-bis(oxidanyl)hexadeca-9,15-dien-11,13-diynoic acid, Acetyl-CoA C-acyltransferase | | Authors: | Lin, C.C, Ko, T.P, Huang, K.F, Yang, Y.L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Integrated omics approach to unveil antifungal bacterial polyynes as acetyl-CoA acetyltransferase inhibitors.
Commun Biol, 5, 2022
|
|
3EGA
 
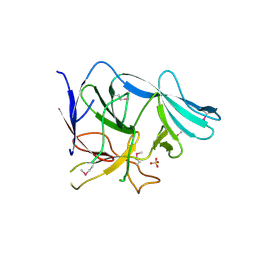 | |
3EGB
 
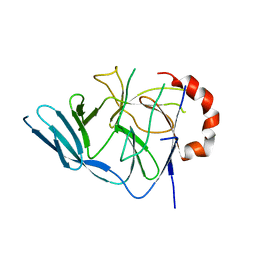 | |
2ZI0
 
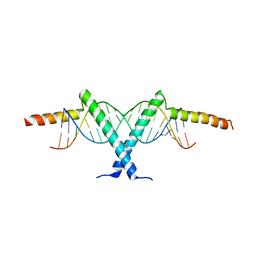 | | Crystal structure of Tav2b/siRNA complex | | Descriptor: | Protein 2b, RNA (5'-D(P*AP*GP*AP*CP*AP*GP*CP*AP*UP*UP*AP*UP*GP*CP*UP*GP*UP*CP*UP*UP*U)-3') | | Authors: | Yuan, Y.A, Chen, H.-Y. | | Deposit date: | 2008-02-12 | | Release date: | 2008-07-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis for RNA-silencing suppression by Tomato aspermy virus protein 2b
Embo Rep., 9, 2008
|
|
7F7N
 
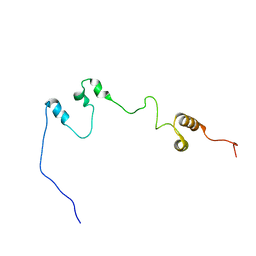 | |
2A4G
 
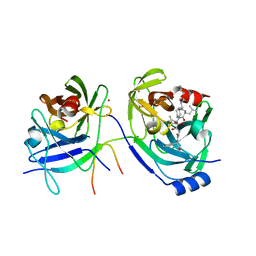 | | Hepatitis C Protease NS3-4A serine protease with Ketoamide Inhibitor SCH225724 Bound | | Descriptor: | ({1-[1-CARBAMOYL-PHENYL-METHYL)-CARBAMOYL]-METHYL}-AMINOOXALYL)-BUTYLCARBAMOYL)-3-METHYL-BUTYLCARBAMOYL)-CYCLOHEXYL-METHYL)-CARBAMIC ACID ISOBUTYL ESTER, NS3 protease/helicase, NS4a peptide, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chan, T.Y, Bennett, F, Bogen, S.L, Chen, K, Gu, H, Hong, L, Jao, E, Liu, Y.T, Lovey, R.G, Parekh, T, Pike, R.E, Pinto, P, Santhanam, B, Venkatraman, S, Vaccaro, H, Wang, H, Yang, X, Zhu, Z, Mckittrick, B, Saksena, A.K, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Ingram, R, Malcolm, B, Prongay, A.J, Yao, N, Marten, B, Madison, V, Kemp, S, Levy, O, Lim-Wilby, M, Tamura, S, Ganguly, A.K. | | Deposit date: | 2005-06-28 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Hepatitis C virus NS3-4a serine protease inhibitors. SAR of P2' moiety with improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A4R
 
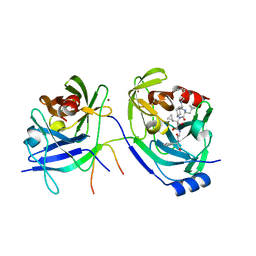 | | HCV NS3 Protease Domain with a Ketoamide Inhibitor Covalently bound. | | Descriptor: | NS3 protease/helicase, Ns4a peptide, ZINC ION, ... | | Authors: | Bogen, S, Saksena, A.K, Arasappan, A, Gu, H, Njoroge, F.G, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hepatitis C Virus NS3-4A serine protease inhibitors: Use of a P2-P1 cyclopropyl alanine combination for improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A4Q
 
 | | HCV NS3 protease with NS4a peptide and a covalently bound macrocyclic ketoamide compound. | | Descriptor: | (2R)-({N-[(3S)-3-({[(3S,6S)-6-CYCLOHEXYL-5,8-DIOXO-4,7-DIAZABICYCLO[14.3.1]ICOSA-1(20),16,18-TRIEN-3-YL]CARBONYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETIC ACID, BETA-MERCAPTOETHANOL, NS3 protease/helicase', ... | | Authors: | Chen, K.X, Njoroge, F.G, Prongay, A, Pichardo, J, Madison, V, Girijavallabhan, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and Biological Activity of Macrocyclic Inhibitors of Hepatitis C Virus (HCV) NS3 Protease
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2GVF
 
 | | HCV NS3-4A protease domain complexed with a macrocyclic ketoamide inhibitor, SCH419021 | | Descriptor: | (6R,8S,11S)-11-CYCLOHEXYL-N-(1-{[(2-{[(1S)-2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO}-2-OXOETHYL)AMINO](OXO)ACETYL}BUTYL)-10,13-DIOXO-2,5-DIOXA-9,12-DIAZATRICYCLO[14.3.1.1~6,9~]HENICOSA-1(20),16,18-TRIENE-8-CARBOXAMIDE, Polyprotein, ZINC ION, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chen, K.X, Venkatraman, S, Parekh, T.N, Gu, H, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V, Girijavallabhan, V. | | Deposit date: | 2006-05-02 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4M8Z
 
 | |
2OIN
 
 | | crystal structure of HCV NS3-4A R155K mutant | | Descriptor: | NS4A peptide, Polyprotein, ZINC ION | | Authors: | Wei, Y. | | Deposit date: | 2007-01-11 | | Release date: | 2007-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: sensitivity to telaprevir (VX-950) and interferon alpha.
J.Biol.Chem., 282, 2007
|
|
2EFJ
 
 | |
