3KP1
 
 | | Crystal structure of ornithine 4,5 aminomutase (Resting State) | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOW
 
 | | Crystal Structure of ornithine 4,5 aminomutase backsoaked complex | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOX
 
 | | Crystal Structure of ornithine 4,5 aminomutase in complex with 2,4-diaminobutyrate (Anaerobic) | | Descriptor: | (2S)-2-amino-4-{[(1Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}butanoic acid, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOZ
 
 | | Crystal Structure of ornithine 4,5 aminomutase in complex with ornithine (Anaerobic) | | Descriptor: | (E)-N~5~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-ornithine, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
6S7E
 
 | | Plant Cysteine Oxidase PCO4 from Arabidopsis thaliana (using PEG 3350 and NaF as precipitants) | | Descriptor: | FE (III) ION, Plant cysteine oxidase 4 | | Authors: | White, M.D, Levy, C.W, Flashman, E, McDonough, M.A. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-22 | | Last modified: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of Arabidopsis thaliana oxygen-sensing plant cysteine oxidases 4 and 5 enable targeted manipulation of their activity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6QVI
 
 | |
6R48
 
 | |
6R46
 
 | |
6GKV
 
 | | Crystal structure of Coclaurine N-Methyltransferase (CNMT) bound to N-methylheliamine and SAH | | Descriptor: | 6,7-dimethoxy-2-methyl-1,2,3,4-tetrahydroisoquinolin-2-ium, Coclaurine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Dunstan, M.S, Levy, C.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-06-06 | | Last modified: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Biocatalytic Scope of Coclaurine N-Methyltransferase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GKY
 
 | |
6GKZ
 
 | |
6FYJ
 
 | | Cytochrome P450 peroxygenase CYP152K6 in complex with Myristic Acid | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fatty-acid peroxygenase, MYRISTIC ACID, ... | | Authors: | Girvan, H.M, Poddar, H, Mclean, K.J, Leys, D, Munro, A.W. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and catalytic properties of the peroxygenase P450 enzyme CYP152K6 from Bacillus methanolicus.
J. Inorg. Biochem., 188, 2018
|
|
6H1T
 
 | | Structure of the BM3 heme domain in complex with clotrimazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
6H1S
 
 | | Structure of the BM3 heme domain in complex with fluconazole | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
6H1L
 
 | | Structure of the BM3 heme domain in complex with tioconazole | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
6H1O
 
 | | Structure of the BM3 heme domain in complex with voriconazole | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.734 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
7NQM
 
 | | Mycobacterium tuberculosis Cytochrome P450 CYP121 in complex with lead compound 10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(2-pyrimidin-4-ylethyl)-1~{H}-indole, CHLORIDE ION, ... | | Authors: | Selvam, I.R. | | Deposit date: | 2021-03-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new strategy for hit generation: Novel in cellulo active inhibitors of CYP121A1 from Mycobacterium tuberculosis via a combined X-ray crystallographic and phenotypic screening approach (XP screen).
Eur.J.Med.Chem., 230, 2022
|
|
7NQN
 
 | | Mycobacterium tuberculosis Cytochrome P450 CYP121 in complex with lead compound 14 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[4-[2-(1~{H}-indol-3-yl)ethyl]pyrimidin-2-yl]morpholine, CHLORIDE ION, ... | | Authors: | Selvam, I.R. | | Deposit date: | 2021-03-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new strategy for hit generation: Novel in cellulo active inhibitors of CYP121A1 from Mycobacterium tuberculosis via a combined X-ray crystallographic and phenotypic screening approach (XP screen).
Eur.J.Med.Chem., 230, 2022
|
|
7NQO
 
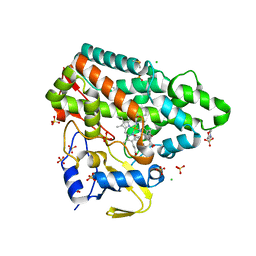 | | Mycobacterium tuberculosis Cytochrome P450 CYP121 in complex with lead compound 21 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[4-[2-(5-bromanyl-1~{H}-indol-3-yl)ethyl]pyrimidin-2-yl]morpholine, CHLORIDE ION, ... | | Authors: | Selvam, I.R. | | Deposit date: | 2021-03-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new strategy for hit generation: Novel in cellulo active inhibitors of CYP121A1 from Mycobacterium tuberculosis via a combined X-ray crystallographic and phenotypic screening approach (XP screen).
Eur.J.Med.Chem., 230, 2022
|
|
8C49
 
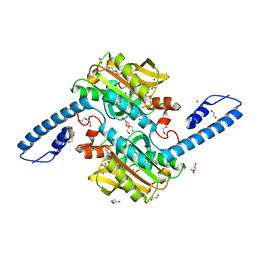 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for 3-Methyl-L-histidine, bound to AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hardy, F.J, Levy, C.W. | | Deposit date: | 2023-01-03 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Engineering mutually orthogonal PylRS/tRNA pairs for dual encoding of functional histidine analogues.
Protein Sci., 32, 2023
|
|
