7D2P
 
 | |
7D28
 
 | |
7D2M
 
 | |
5SV6
 
 | |
5DBT
 
 | |
3U0R
 
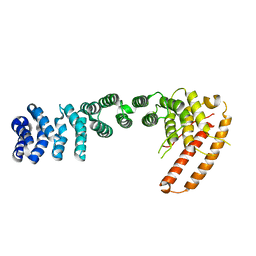 | | Helical repeat structure of apoptosis inhibitor 5 reveals protein-protein interaction modules | | Descriptor: | Apoptosis inhibitor 5 | | Authors: | Han, B.G, Kim, K.H, Jeong, K.C, Cho, J.W, Noh, K.H, Kim, T.W, Yoon, H.J, Suh, S.W, Lee, S.H, Lee, B.I. | | Deposit date: | 2011-09-29 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Helical repeat structure of apoptosis inhibitor 5 reveals protein-protein interaction modules.
J.Biol.Chem., 287, 2012
|
|
5DBU
 
 | |
7YPH
 
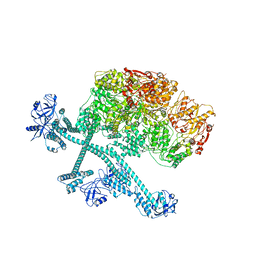 | | Open-spiral pentamer of the substrate-free Lon protease with a Y224S mutation | | Descriptor: | Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPJ
 
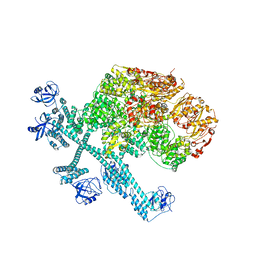 | | Spiral pentamer of the substrate-free Lon protease with a S678A mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPK
 
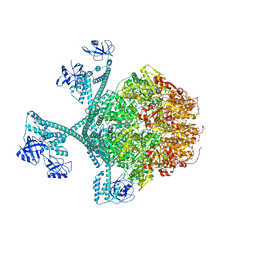 | | Close-ring hexamer of the substrate-bound Lon protease with an S678A mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, alpha-S1-casein | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPI
 
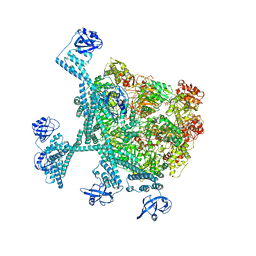 | | Spiral hexamer of the substrate-free Lon protease with a Y224S mutation | | Descriptor: | Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
1ZM1
 
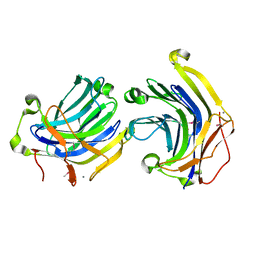 | | Crystal structures of complex F. succinogenes 1,3-1,4-beta-D-glucanase and beta-1,3-1,4-cellotriose | | Descriptor: | Beta-glucanase, CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Tsai, L.C, Shyur, L.F, Cheng, Y.S, Lee, S.H. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase in complex with beta-1,3-1,4-cellotriose
J.Mol.Biol., 354, 2005
|
|
6AFP
 
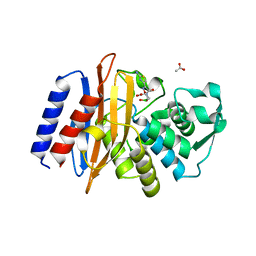 | | Crystal structure of class A b-lactamase, PenL, variant Asn136Asp, from Burkholderia thailandensis, in complex with ceftazidime-like boronic acid | | Descriptor: | ACETATE ION, Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Cao, T.-P, Choi, J.M, Lee, S.H. | | Deposit date: | 2018-08-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Non-catalytic-Region Mutations Conferring Transition of Class A beta-Lactamases Into ESBLs.
Front Mol Biosci, 2020
|
|
6AFO
 
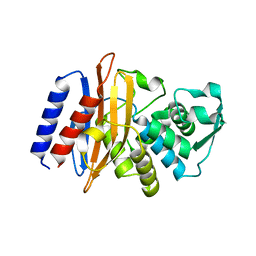 | |
6AFM
 
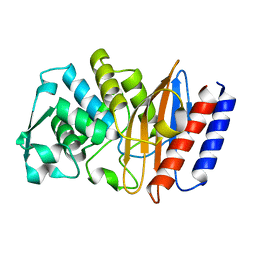 | |
6AFN
 
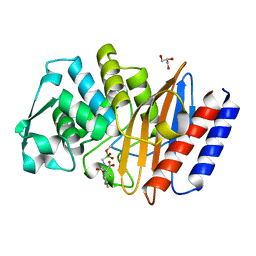 | | Crystal structure of class A b-lactamase, PenL, variant Cys69Tyr, from Burkholderia thailandensis, in complex with ceftazidime-like boronic acid | | Descriptor: | Beta-lactamase, GLYCEROL, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Cao, T.-P, Choi, J.M, Lee, S.H. | | Deposit date: | 2018-08-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Non-catalytic-Region Mutations Conferring Transition of Class A beta-Lactamases Into ESBLs.
Front Mol Biosci, 2020
|
|
6J4N
 
 | | Structure of papua new guinea MBL-1(PNGM-1) native | | Descriptor: | Metallo-beta-lactamases PNGM-1, ZINC ION | | Authors: | Hong, M.K, Park, K.S, Jeon, J.H, Lee, J.H, Park, Y.S, Lee, S.H, Kang, L.W. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The novel metallo-beta-lactamase PNGM-1 from a deep-sea sediment metagenome: crystallization and X-ray crystallographic analysis.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6LTQ
 
 | | Crystal structure of pyrrolidone carboxyl peptidase from thermophilic keratin degrading bacterium Fervidobacterium islandicum AW-1 (FiPcp) | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Pyroglutamyl-peptidase I | | Authors: | Dhanasingh, I, Jin, H.S, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-01-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of oxidized pyrrolidone carboxypeptidase from Fervidobacterium islandicum AW-1 reveals unique structural features for thermostability and keratinolysis.
Biochem.Biophys.Res.Commun., 540, 2021
|
|
7C1C
 
 | |
7C18
 
 | |
7C1A
 
 | |
7C90
 
 | | Crystal structure of Cytochrome CL from the marine methylotrophic bacterium Methylophaga aminisulfidivorans MPT (Ma-CytcL) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Cytochrome c, ... | | Authors: | Ghosh, S, Dhanasingh, I, Lee, S.H. | | Deposit date: | 2020-06-04 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of CytochromecLfrom the Aquatic Methylotrophic BacteriumMethylophaga aminisulfidivoransMPT.
J Microbiol Biotechnol., 30, 2020
|
|
7CVX
 
 | |
7CMY
 
 | | Isocitrate lyase from Bacillus cereus ATCC 14579 in complex with Magnessium ion, glyoxylate, and succinate | | Descriptor: | GLYOXYLIC ACID, Isocitrate lyase, MAGNESIUM ION, ... | | Authors: | Kim, K, Ki, D, Lee, S.H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isocitrate lyase from Bacillus cereus ATCC 14579 in complex with Magnessium ion, glyoxylate, and succinate
To Be Published
|
|
7CMX
 
 | |
