3KTI
 
 | | Structure of ClpP in complex with ADEP1 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ATP-dependent Clp protease proteolytic subunit, Acyldepsipeptide 1, ... | | Authors: | Lee, B.-G, Brotz-Oesterhelt, H, Song, H.K. | | Deposit date: | 2009-11-25 | | Release date: | 2010-03-23 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KTK
 
 | |
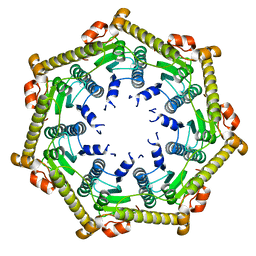
3KTH
 
 | |
3KTG
 
 | |
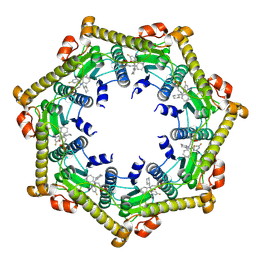
3KTJ
 
 | |
3TT7
 
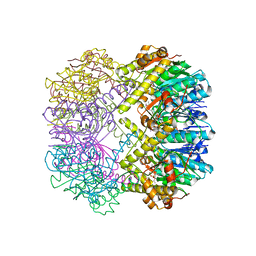 | |
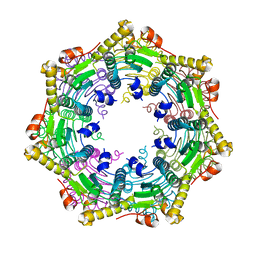
3TT6
 
 | |
7OGT
 
 | | Folded elbow of cohesin | | Descriptor: | Structural maintenance of chromosomes protein 1, Structural maintenance of chromosomes protein 3 | | Authors: | Lee, B.-G, Gonzalez Llamazares, A, Collier, J, Patele, N.J, Nasmyth, K.A, Lowe, J. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Folding of cohesin's coiled coil is important for Scc2/4-induced association with chromosomes.
Elife, 10, 2021
|
|
6ZZ6
 
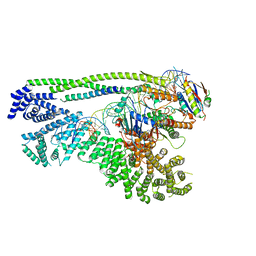 | | Cryo-EM structure of S.cerevisiae cohesin-Scc2-DNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (34-MER), MAGNESIUM ION, ... | | Authors: | Lee, B.-G, Gonzalez Llamazares, A, Collier, J, Nasmyth, K.A, Lowe, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transport of DNA within cohesin involves clamping on top of engaged heads by Scc2 and entrapment within the ring by Scc3.
Elife, 9, 2020
|
|
4FBC
 
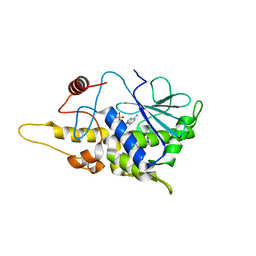 | | Structure of mutant RIP from barley seeds in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FB9
 
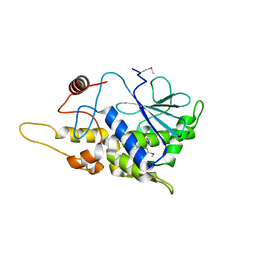 | | Structure of mutant RIP from barley seeds | | Descriptor: | Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6YVV
 
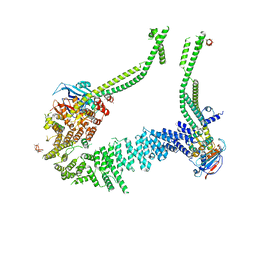 | | Condensin complex from S.cerevisiae ATP-free apo bridged state | | Descriptor: | Condensin complex subunit 1,Ycs4, Condensin complex subunit 2,Brn1, Structural maintenance of chromosomes protein 2,Structural maintenance of chromosomes protein 2, ... | | Authors: | Lee, B.-G, Cawood, C, Gutierrez-Escribano, P, Nakane, T, Merkel, F, Hassler, M, Haering, C.H, Aragon, L, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6YVU
 
 | | Condensin complex from S.cerevisiae ATP-free apo non-engaged state | | Descriptor: | Condensin complex subunit 1,Condensin complex subunit 1,Ycs4, Condensin complex subunit 2,Condensin complex subunit 2,Brn1, Structural maintenance of chromosomes protein 2,Structural maintenance of chromosomes protein 2,Smc2, ... | | Authors: | Lee, B.-G, Cawood, C, Gutierrez-Escribano, P, Nakane, T, Merkel, F, Hassler, M, Aragon, L, Haering, C.H, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5F0N
 
 | | Cohesin subunit Pds5 | | Descriptor: | cohesin subunit Pds5,cohesin subunit Pds5, cohesin subunit Pds5,KLTH0D07062p,cohesin subunit Pds5,cohesin subunit Pds5, cohesin subunit Pds5 | | Authors: | Lee, B.-G, Jansma, M, Nasmyth, K, Lowe, J. | | Deposit date: | 2015-11-27 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Cohesin Gatekeeper Pds5 and in Complex with Kleisin Scc1.
Cell Rep, 14, 2016
|
|
5F0O
 
 | | Cohesin subunit Pds5 in complex with Scc1 | | Descriptor: | KLTH0G16610p, cohesin subunit Pds5, KLTH0D07062p,KLTH0D07062p,KLTH0D07062p,cohesin subunit Pds5, ... | | Authors: | Lee, B.-G, Jansma, M, Nasmyth, K, Lowe, J. | | Deposit date: | 2015-11-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of the Cohesin Gatekeeper Pds5 and in Complex with Kleisin Scc1.
Cell Rep, 14, 2016
|
|
4FBA
 
 | | Structure of mutant RIP from barley seeds in complex with adenine | | Descriptor: | ADENINE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FBB
 
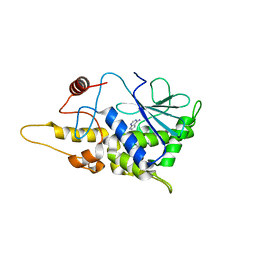 | | Structure of mutant RIP from barley seeds in complex with adenine (AMP-incubated) | | Descriptor: | ADENINE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FBH
 
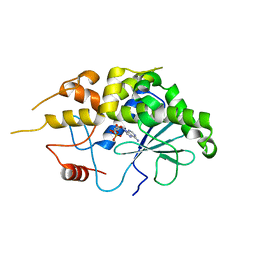 | | Structure of RIP from barley seeds | | Descriptor: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7P80
 
 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compressed state) | | Descriptor: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7P81
 
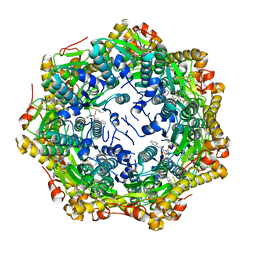 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compact state) | | Descriptor: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7Q2X
 
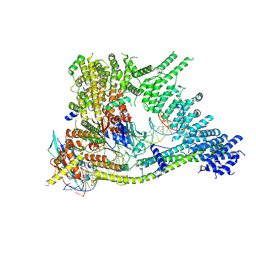 | | Cryo-EM structure of clamped S.cerevisiae condensin-DNA complex (Form I) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Condensin complex subunit 1, ... | | Authors: | Lee, B.-G, Rhodes, J, Lowe, J. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Clamping of DNA shuts the condensin neck gate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q2Y
 
 | | Cryo-EM structure of clamped S.cerevisiae condensin-DNA complex (form II) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Condensin complex subunit 1, ... | | Authors: | Lee, B.-G, Rhodes, J, Lowe, J. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-23 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Clamping of DNA shuts the condensin neck gate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q2Z
 
 | |
5B62
 
 | | Crystal structure of N-terminal amidase with Asn-Glu-Ala peptide | | Descriptor: | ASN-GLU-ALA, Nta1p | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.042 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5HYY
 
 | |
