2V8Z
 
 | | Crystal Structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 | | 分子名称: | YAGE | | 著者 | Manicka, S, Peleg, Y, Unger, T, Albeck, S, Dym, O, Greenblatt, H.M, Bourenkov, G, Lamzin, V, Krishnaswamy, S, Sussman, J.L. | | 登録日 | 2007-08-16 | | 公開日 | 2008-03-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Yage, a Putative Dhdps Like Protein from Escherichia Coli K12.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2V9D
 
 | | Crystal Structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 | | 分子名称: | YAGE | | 著者 | Manicka, S, Peleg, Y, Unger, T, Albeck, S, Dym, O, Greenblatt, H.M, Bourenkov, G, Lamzin, V, Krishnaswamy, S, Sussman, J.L. | | 登録日 | 2007-08-23 | | 公開日 | 2008-03-04 | | 最終更新日 | 2019-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal Structure of Yage, a Putative Dhdps Like Protein from Escherichia Coli K12.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
1EJG
 
 | | CRAMBIN AT ULTRA-HIGH RESOLUTION: VALENCE ELECTRON DENSITY. | | 分子名称: | CRAMBIN (PRO22,SER22/LEU25,ILE25) | | 著者 | Jelsch, C, Teeter, M.M, Lamzin, V, Pichon-Lesme, V, Blessing, B, Lecomte, C. | | 登録日 | 2000-03-02 | | 公開日 | 2000-04-05 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (0.54 Å) | | 主引用文献 | Accurate protein crystallography at ultra-high resolution: valence electron distribution in crambin.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EGP
 
 | |
1CEX
 
 | | STRUCTURE OF CUTINASE | | 分子名称: | CUTINASE | | 著者 | Longhi, S, Czjzek, M, Lamzin, V, Nicolas, A, Cambillau, C. | | 登録日 | 1997-02-18 | | 公開日 | 1997-08-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Atomic resolution (1.0 A) crystal structure of Fusarium solani cutinase: stereochemical analysis.
J.Mol.Biol., 268, 1997
|
|
1CKU
 
 | | AB INITIO SOLUTION AND REFINEMENT OF TWO HIGH POTENTIAL IRON PROTEIN STRUCTURES AT ATOMIC RESOLUTION | | 分子名称: | IRON/SULFUR CLUSTER, PROTEIN (HIPIP) | | 著者 | Parisini, E, Capozzi, F, Lubini, P, Lamzin, V, Luchinat, C, Sheldrick, G.M. | | 登録日 | 1999-04-24 | | 公開日 | 1999-05-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Ab initio solution and refinement of two high-potential iron protein structures at atomic resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1I6U
 
 | | RNA-PROTEIN INTERACTIONS: THE CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN S8/RRNA COMPLEX FROM METHANOCOCCUS JANNASCHII | | 分子名称: | 16S RRNA FRAGMENT, 30S RIBOSOMAL PROTEIN S8P, SULFATE ION | | 著者 | Tishchenko, S, Nikulin, A, Fomenkova, N, Nevskaya, N, Nikonov, O, Dumas, P, Moine, H, Ehresmann, B, Ehresmann, C, Piendl, W, Lamzin, V, Garber, M, Nikonov, S. | | 登録日 | 2001-03-05 | | 公開日 | 2001-08-03 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Detailed analysis of RNA-protein interactions within the ribosomal protein S8-rRNA complex from the archaeon Methanococcus jannaschii.
J.Mol.Biol., 311, 2001
|
|
1KWF
 
 | | Atomic Resolution Structure of an Inverting Glycosidase in Complex with Substrate | | 分子名称: | Endoglucanase A, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Guerin, D.M.A, Lascombe, M.-B, Costabel, M, Souchon, H, Lamzin, V, Beguin, P, Alzari, P.M. | | 登録日 | 2002-01-29 | | 公開日 | 2002-03-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (0.94 Å) | | 主引用文献 | Atomic (0.94 A) resolution structure of an inverting glycosidase in complex with substrate.
J.Mol.Biol., 316, 2002
|
|
7QOR
 
 | | Structure of beta-lactamase TEM-171 | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | 著者 | Hakanpaa, J, Petrova, T, Samygina, V.R, Chojnowski, G, Lamzin, V, Egorov, A.M. | | 登録日 | 2021-12-28 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Crystal structures of the molecular class A beta-lactamase TEM-171 and its complexes with tazobactam.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4U4M
 
 | | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate | | 分子名称: | 1,2-ETHANEDIOL, PYRUVIC ACID, UREA, ... | | 著者 | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | 登録日 | 2014-07-24 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate
To be published
|
|
4LZT
 
 | | ATOMIC RESOLUTION REFINEMENT OF TRICLINIC HEW LYSOZYME AT 295K | | 分子名称: | LYSOZYME, NITRATE ION | | 著者 | Walsh, M.A, Schneider, T, Sieker, L.C, Dauter, Z, Lamzin, V, Wilson, K.S. | | 登録日 | 1997-03-31 | | 公開日 | 1998-04-01 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Refinement of triclinic hen egg-white lysozyme at atomic resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3LZT
 
 | | REFINEMENT OF TRICLINIC LYSOZYME AT ATOMIC RESOLUTION | | 分子名称: | ACETATE ION, LYSOZYME, NITRATE ION | | 著者 | Walsh, M.A, Schneider, T, Sieker, L.C, Dauter, Z, Lamzin, V, Wilson, K.S. | | 登録日 | 1997-03-23 | | 公開日 | 1998-03-25 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (0.925 Å) | | 主引用文献 | Refinement of triclinic hen egg-white lysozyme at atomic resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
4PTN
 
 | | Crystal Structure of YagE, a KDG aldolase protein in complex with Magnesium cation coordinated L-glyceraldehyde | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, L-glyceraldehyde, ... | | 著者 | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | 登録日 | 2014-03-11 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal structure of YagE, a putative DHDPS-like protein from Escherichia coli K12.
Proteins, 71, 2008
|
|
4OE7
 
 | | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal | | 分子名称: | (4R)-4-hydroxy-2,5-dioxopentanoic acid, (4S)-4-hydroxy-2,5-dioxopentanoic acid, 1,2-ETHANEDIOL, ... | | 著者 | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | 登録日 | 2014-01-12 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal
To be Published
|
|
4ONV
 
 | | Crystal structure of YagE, a KDG aldolase protein in complex with 2-Keto-3-deoxy gluconate | | 分子名称: | 1,2-ETHANEDIOL, 2-KETO-3-DEOXYGLUCONATE, GLYCEROL, ... | | 著者 | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | 登録日 | 2014-01-29 | | 公開日 | 2015-01-14 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Crystal structure of YagE, a KDG aldolase protein in complex with 2-Keto-3-deoxy gluconate
To be Published
|
|
1AGY
 
 | |
1B0Y
 
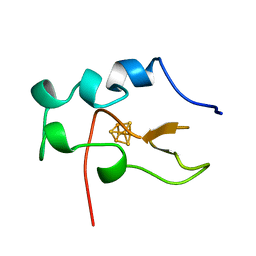 | |
2PVB
 
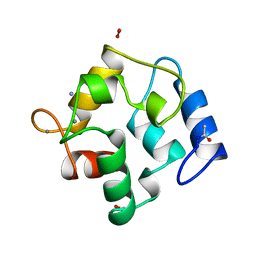 | |
1EE2
 
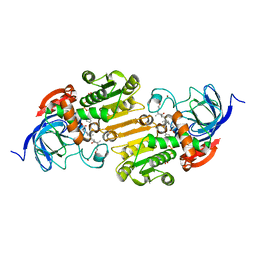 | |
1FH2
 
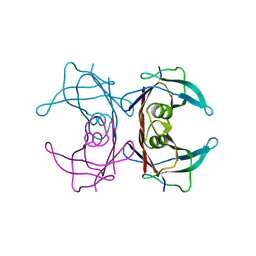 | |
1FHN
 
 | |
1F86
 
 | | TRANSTHYRETIN THR119MET PROTEIN STABILISATION | | 分子名称: | 3,5,3',5'-TETRAIODO-L-THYRONINE, TRANSTHYRETIN THR119MET VARIANT | | 著者 | Sebastiao, M.P. | | 登録日 | 2000-06-29 | | 公開日 | 2001-06-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Transthyretin stability as a key factor in amyloidogenesis: X-ray analysis at atomic resolution.
J.Mol.Biol., 306, 2001
|
|
1HBU
 
 | |
1HBO
 
 | | METHYL-COENZYME M REDUCTASE MCR-RED1-SILENT | | 分子名称: | 1-THIOETHANESULFONIC ACID, CHLORIDE ION, Coenzyme B, ... | | 著者 | Grabarse, W. | | 登録日 | 2001-04-20 | | 公開日 | 2001-08-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | On the Mechanism of Biological Methane Formation: Structural Evidence for Conformational Changes in Methyl-Coenzyme M Reductase Upon Substrate Binding
J.Mol.Biol., 309, 2001
|
|
1HBM
 
 | | METHYL-COENZYME M REDUCTASE ENZYME PRODUCT COMPLEX | | 分子名称: | CHLORIDE ION, FACTOR 430, GLYCEROL, ... | | 著者 | Ermler, U, Grabarse, W. | | 登録日 | 2001-04-20 | | 公開日 | 2001-08-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | On the Mechanism of Biological Methane Formation: Structural Evidence for Conformational Changes in Methyl-Coenzyme M Reductase Upon Substrate Binding
J.Mol.Biol., 309, 2001
|
|
