8R96
 
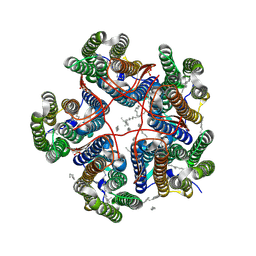 | |
8R97
 
 | | Crystal structure of the cryorhodopsin CryoR2 at pH 4.6, type B crystals, non-illuminated state | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Lamm, G.H.U, Marin, E. | | Deposit date: | 2023-11-30 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8R98
 
 | | Crystal structure of the cryorhodopsin CryoR2 at pH 4.6, type B crystals, illuminated state | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Lamm, G.H.U, Marin, E. | | Deposit date: | 2023-11-30 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8R0P
 
 | | Cryo-EM structure of the microbial rhodopsin CryoR2 at pH 8.0 in detergent | | Descriptor: | EICOSANE, RETINAL, microbial rhodopsin CryoR2 | | Authors: | Kovalev, K, Marin, E, Stetsenko, A, Guskov, A, Lamm, G.H.U. | | Deposit date: | 2023-10-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8R0L
 
 | | Cryo-EM structure of the microbial rhodopsin CryoR1 at pH 8.0 in nanodisc | | Descriptor: | EICOSANE, RETINAL, Rhodopsin | | Authors: | Kovalev, K, Marin, E, Stetsenko, A, Guskov, A, Lamm, G.H.U. | | Deposit date: | 2023-10-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8R0K
 
 | | Cryo-EM structure of the microbial rhodopsin CryoR1 at pH 4.3 in detergent | | Descriptor: | EICOSANE, RETINAL, Rhodopsin | | Authors: | Kovalev, K, Marin, E, Stetsenko, A, Guskov, A, Lamm, G.H.U. | | Deposit date: | 2023-10-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8R0N
 
 | | Cryo-EM structure of the microbial rhodopsin CryoR1 at pH 10.5 in detergent in the ground state | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Marin, E, Stetsenko, A, Guskov, A, Lamm, G.H.U. | | Deposit date: | 2023-10-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8R0M
 
 | | Cryo-EM structure of the microbial rhodopsin CryoR1 at pH 8.0 in detergent | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Marin, E, Stetsenko, A, Guskov, A, Lamm, G.H.U. | | Deposit date: | 2023-10-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8R0O
 
 | | Cryo-EM structure of the microbial rhodopsin CryoR1 at pH 10.5 in detergent in the M state | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, RETINAL, Rhodopsin | | Authors: | Kovalev, K, Marin, E, Stetsenko, A, Guskov, A, Lamm, G.H.U. | | Deposit date: | 2023-10-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8QLE
 
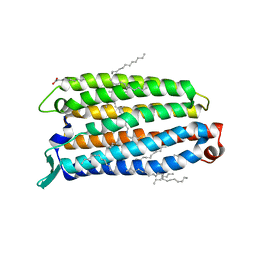 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 4.6 | | Descriptor: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | Deposit date: | 2023-09-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QLF
 
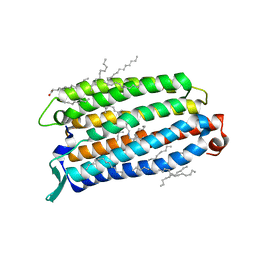 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 8.8 | | Descriptor: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | Deposit date: | 2023-09-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QQZ
 
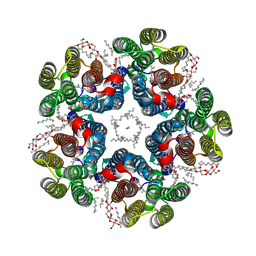 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 8.0 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QR0
 
 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 4.3 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
9IN8
 
 | | True-atomic resolution crystal structure of the open state of the viral channelrhodopsin OLPVR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, EICOSANE, ... | | Authors: | Bukhdruker, S, Zabelskii, D, Astashkin, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2024-07-05 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Ion-conducting and gating molecular mechanisms of channelrhodopsin revealed by true-atomic-resolution structures of open and closed states.
Nat.Struct.Mol.Biol., 2025
|
|
9IN9
 
 | | Structure of the viral channelrhodopsin OLPVR1 at low pH obtained by soaking | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, EICOSANE, ... | | Authors: | Bukhdruker, S, Zabelskii, D, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-07-05 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Ion-conducting and gating molecular mechanisms of channelrhodopsin revealed by true-atomic-resolution structures of open and closed states.
Nat.Struct.Mol.Biol., 2025
|
|
9IN7
 
 | | True-atomic resolution crystal structure of the closed state of the viral channelrhodopsin OLPVR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, EICOSANE, ... | | Authors: | Bukhdruker, S, Zabelskii, D, Astashkin, R, Gordeliy, V. | | Deposit date: | 2024-07-05 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Ion-conducting and gating molecular mechanisms of channelrhodopsin revealed by true-atomic-resolution structures of open and closed states.
Nat.Struct.Mol.Biol., 2025
|
|
