2MBS
 
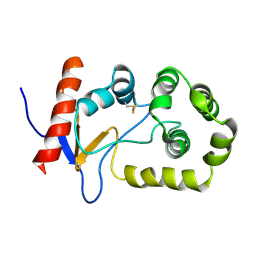 | | NMR solution structure of oxidized KpDsbA | | 分子名称: | Thiol:disulfide interchange protein | | 著者 | Kurth, F, Rimmer, K, Premkumar, L, Mohanty, B, Duprez, W, Halili, M.A, Shouldice, S.R, Heras, B, Fairlie, D.P, Scanlon, M.J, Martin, J.L. | | 登録日 | 2013-08-03 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comparative Sequence, Structure and Redox Analyses of Klebsiella pneumoniae DsbA Show That Anti-Virulence Target DsbA Enzymes Fall into Distinct Classes.
Plos One, 8, 2013
|
|
4MCU
 
 | |
4OCF
 
 | |
4OCE
 
 | |
4OD7
 
 | |
6NEN
 
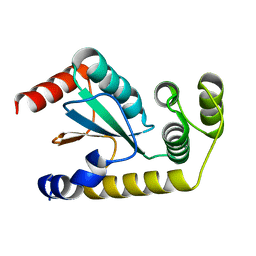 | | Catalytic domain of Proteus mirabilis ScsC | | 分子名称: | Copper resistance protein | | 著者 | Kurth, F, Furlong, E.J, Premkumar, L, Martin, J.L. | | 登録日 | 2018-12-17 | | 公開日 | 2019-03-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.151 Å) | | 主引用文献 | Engineered variants provide new insight into the structural properties important for activity of the highly dynamic, trimeric protein disulfide isomerase ScsC from Proteus mirabilis.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4XVW
 
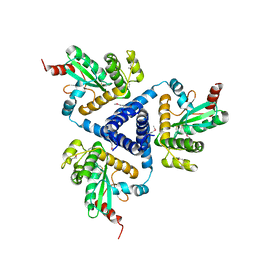 | |
5ID4
 
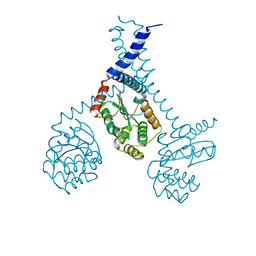 | |
6C29
 
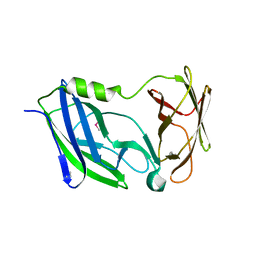 | | Crystal structure of the N-terminal periplasmic domain of ScsB from Proteus mirabilis | | 分子名称: | Putative metal resistance protein | | 著者 | Furlong, E.J, Choudhury, H.G, Kurth, F, Martin, J.L. | | 登録日 | 2018-01-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (1.538 Å) | | 主引用文献 | Disulfide isomerase activity of the dynamic, trimericProteus mirabilisScsC protein is primed by the tandem immunoglobulin-fold domain of ScsB.
J. Biol. Chem., 293, 2018
|
|
5IDR
 
 | |
3FBI
 
 | | Structure of the Mediator submodule Med7N/31 | | 分子名称: | Mediator of RNA polymerase II transcription subunit 31, Mediator of RNA polymerase II transcription subunit 7 | | 著者 | Koschubs, T, Seizl, M, Lariviere, L, Kurth, F, Baumli, S, Martin, D.E, Cramer, P. | | 登録日 | 2008-11-19 | | 公開日 | 2008-12-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Identification, structure, and functional requirement of the Mediator submodule Med7N/31
Embo J., 28, 2009
|
|
3FBN
 
 | | Structure of the Mediator submodule Med7N/31 | | 分子名称: | Mediator of RNA polymerase II transcription subunit 31, Mediator of RNA polymerase II transcription subunit 7 | | 著者 | Koschubs, T, Seizl, M, Lariviere, L, Kurth, F, Baumli, S, Martin, D.E, Cramer, P. | | 登録日 | 2008-11-19 | | 公開日 | 2008-12-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.007 Å) | | 主引用文献 | Identification, structure, and functional requirement of the Mediator submodule Med7N/31
Embo J., 28, 2009
|
|
4H61
 
 | | Structure of the Schizosaccharomyces pombe Mediator subunit Med6 | | 分子名称: | Mediator of RNA polymerase II transcription subunit 6 | | 著者 | Lariviere, L, Plaschka, C, Seizl, M, Wenzeck, L, Kurth, F, Cramer, P. | | 登録日 | 2012-09-19 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of the Mediator head module.
Nature, 492, 2012
|
|
4H63
 
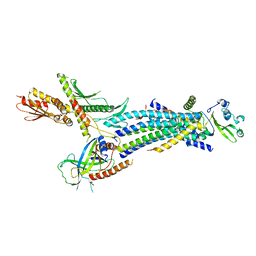 | | Structure of the Schizosaccharomyces pombe Mediator head module | | 分子名称: | Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, Mediator of RNA polymerase II transcription subunit 18, ... | | 著者 | Lariviere, L, Plaschka, C, Seizl, M, Wenzeck, L, Kurth, F, Cramer, P. | | 登録日 | 2012-09-19 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structure of the Mediator head module.
Nature, 492, 2012
|
|
4H62
 
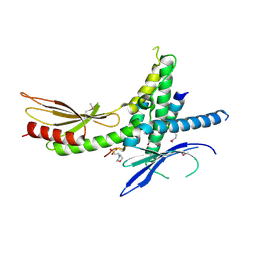 | | Structure of the Saccharomyces cerevisiae Mediator subcomplex Med17C/Med11C/Med22C | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, ... | | 著者 | Lariviere, L, Plaschka, C, Seizl, M, Wenzeck, L, Kurth, F, Cramer, P. | | 登録日 | 2012-09-19 | | 公開日 | 2012-10-31 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of the Mediator head module.
Nature, 492, 2012
|
|
4MLY
 
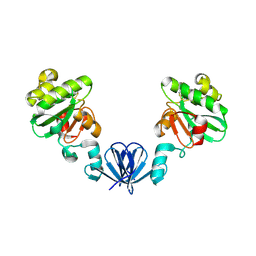 | | Disulfide isomerase from multidrug resistance IncA/C related integrative and conjugative elements in oxidized state (P21 space group) | | 分子名称: | 1,3-BUTANEDIOL, DsbP | | 著者 | Premkumar, L, Kurth, F, Neyer, S, Martin, J.L. | | 登録日 | 2013-09-06 | | 公開日 | 2013-12-11 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.207 Å) | | 主引用文献 | The Multidrug Resistance IncA/C Transferable Plasmid Encodes a Novel Domain-swapped Dimeric Protein-disulfide Isomerase.
J.Biol.Chem., 289, 2014
|
|
4ML6
 
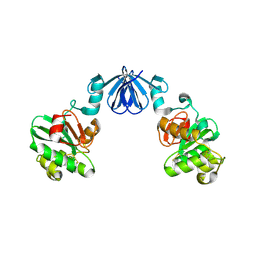 | |
4ML1
 
 | |
6MHH
 
 | |
6XKQ
 
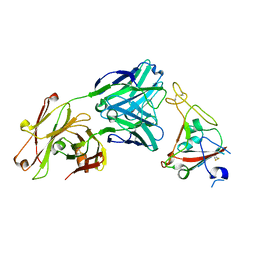 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-250 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-250 Heavy Chain, CV07-250 Light Chain, ... | | 著者 | Yuan, M, Liu, H, Zhu, X, Wu, N.C, Wilson, I.A. | | 登録日 | 2020-06-26 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
6XKP
 
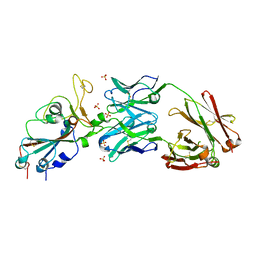 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-270 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-270 Heavy Chain, CV07-270 Light Chain, ... | | 著者 | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | 登録日 | 2020-06-26 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
4GXZ
 
 | | Crystal structure of a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium | | 分子名称: | Suppression of copper sensitivity protein | | 著者 | Shepherd, M, Heras, B, King, G.J, Argente, M.P, Achard, M.E.S, King, N.P, McEwan, A.G, Schembri, M.A. | | 登録日 | 2012-09-04 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural and functional characterization of ScsC, a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium
Antioxid Redox Signal, 19, 2013
|
|
4K6X
 
 | | Crystal structure of disulfide oxidoreductase from Mycobacterium tuberculosis | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Disulfide oxidoreductase | | 著者 | Premkumar, L, Martin, J.L. | | 登録日 | 2013-04-16 | | 公開日 | 2013-10-02 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.972 Å) | | 主引用文献 | Rv2969c, essential for optimal growth in Mycobacterium tuberculosis, is a DsbA-like enzyme that interacts with VKOR-derived peptides and has atypical features of DsbA-like disulfide oxidases.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4P3Y
 
 | |
