1WU8
 
 | |
1WY0
 
 | |
1WY1
 
 | |
1WXD
 
 | |
1X1Q
 
 | |
1WPY
 
 | |
1WWZ
 
 | |
1WZN
 
 | |
2DM9
 
 | |
2DMA
 
 | |
2CVZ
 
 | |
2DCT
 
 | | Crystal structure of the TT1209 from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, SODIUM ION, hypothetical protein TTHA0104 | | Authors: | Asada, Y, Sugahara, M, Shimizu, K, Yamamoto, H, Shimada, H, Nakamoto, T, Ono, N, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-12 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the TT1209 from Thermus thermophilus HB8
To be Published
|
|
2EL3
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L242M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Asada, Y, Matsuura, Y, Ono, N, Shimada, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-10-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L242M)
To be Published
|
|
2EKZ
 
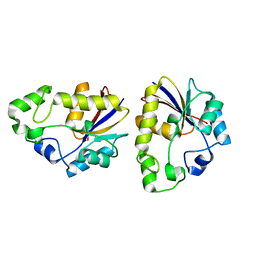 | | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L52M) | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase | | Authors: | Asada, Y, Matsuura, Y, Ono, N, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-10-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L52M)
To be Published
|
|
2EL1
 
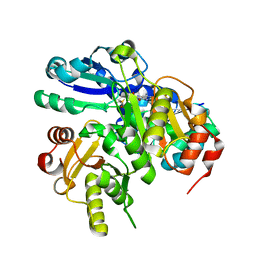 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L44M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Asada, Y, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-10-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L44M)
To be Published
|
|
2EJZ
 
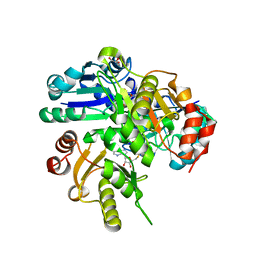 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (Y11M) | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Asada, Y, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (Y11M)
To be Published
|
|
2EL0
 
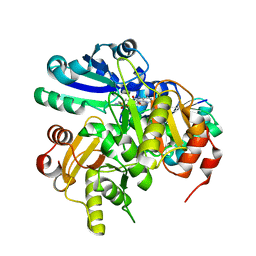 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L21M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, diphthine synthase | | Authors: | Asada, Y, Matsuura, Y, Ono, N, Shimada, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-10-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L21M)
To be Published
|
|
2EOA
 
 | | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (W85H) | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase | | Authors: | Asada, Y, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (W85H)
To be Published
|
|
1ZJJ
 
 | |
1YYA
 
 | |
1ZZG
 
 | |
2CQZ
 
 | |
2CY0
 
 | |
2CSU
 
 | |
2CU2
 
 | |
