2DV7
 
 | | Crystal structure of Lys187 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Saraboji, K, Malathy Sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-28 | | Release date: | 2007-01-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2DSI
 
 | | Crystal structure of Glu171 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Saraboji, K, Malathy Sony, S.M, Ponnuswamy, M.N, Kumarevel, T.S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-06-30 | | Release date: | 2006-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2E15
 
 | |
2E66
 
 | | Crystal Structure Of CutA1 From Pyrococcus Horikoshii OT3, Mutation D60A | | Descriptor: | CHLORIDE ION, Divalent-cation tolerance protein cutA, SODIUM ION | | Authors: | Bagautdinov, B, Sawano, M, Bagautdinova, S, Yutani, K, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-25 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the hyper-thermostability of CutA1
To be Published
|
|
2EEN
 
 | |
2EGL
 
 | | Crystal structure of Glu171 to Lys mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-01 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2EHL
 
 | |
2E8S
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION | | Authors: | Asada, Y, Shimada, H, Taketa, M, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-23 | | Release date: | 2007-07-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2EGS
 
 | | Crystal structure of Leu261 to Met mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-02 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2EJA
 
 | |
2EKA
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L202M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Asada, Y, Matsuura, Y, Kageyama, Y, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L202M)
To be Published
|
|
2EK4
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L8M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Asada, Y, Shimada, H, Taketa, M, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L8M)
To be Published
|
|
2EQ5
 
 | |
2DXW
 
 | |
2DZC
 
 | | Crystal Structure Of Biotin Protein Ligase From Pyrococcus Horikoshii, Mutation R48A | | Descriptor: | biotin--[acetyl-CoA-carboxylase] ligase | | Authors: | Bagautdinov, B, Taketa, M, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-27 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate
J.Biol.Chem., 283, 2008
|
|
2E5F
 
 | |
2E65
 
 | |
2E7R
 
 | | Crystal structure of Glu54 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-12 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of diphthine synthase from Pyrococcus horikoshii OT3
To be Published
|
|
2E41
 
 | | Crystal Structure Of Biotin Protein Ligase From Pyrococcus Horikoshii Complexed with the Reaction Product Analog Biotinol-5'-AMP, Mutations R48A and K111A | | Descriptor: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, biotin--[acetyl-CoA-carboxylase] ligase | | Authors: | Bagautdinov, B, Matsuura, Y, Bagautdinova, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-01 | | Release date: | 2007-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate
J.Biol.Chem., 283, 2008
|
|
2EER
 
 | |
2EHC
 
 | |
2EJG
 
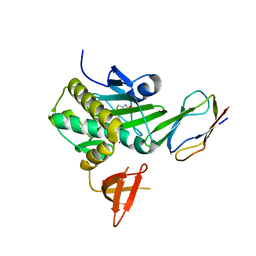 | | Crystal Structure Of The Biotin Protein Ligase (Mutation R48A) and Biotin Carboxyl Carrier Protein Complex From Pyrococcus Horikoshii OT3 | | Descriptor: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain, 235aa long hypothetical biotin--[acetyl-CoA-carboxylase] ligase, ADENOSINE, ... | | Authors: | Bagautdinov, B, Matsuura, Y, Bagautdinova, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate
J.Biol.Chem., 283, 2008
|
|
2EKC
 
 | |
2EJC
 
 | |
2EL2
 
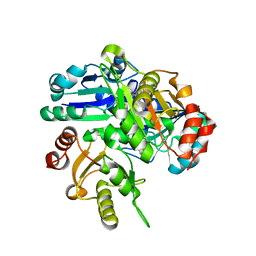 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L185M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Asada, Y, Tanaka, Y, Pampa, K, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-10-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L185M)
To be Published
|
|
