1YYN
 
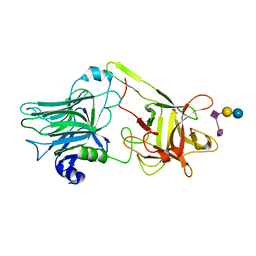 | | A common binding site for disialyllactose and a tri-peptide in the C-fragment of tetanus neurotoxin | | 分子名称: | N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-alpha-D-galactopyranose-(1-4)-beta-D-glucopyranose, Tetanus toxin | | 著者 | Seetharaman, J, Eswaramoorthy, S, Kumaran, D, Swaminathan, S. | | 登録日 | 2005-02-25 | | 公開日 | 2005-03-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Common binding site for disialyllactose and tri-peptide in C-fragment of tetanus neurotoxin
Proteins, 61, 2005
|
|
2QVC
 
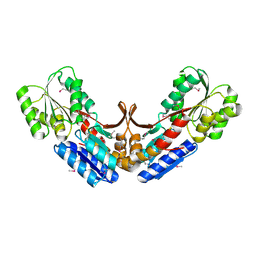 | | Crystal structure of a periplasmic sugar ABC transporter from Thermotoga maritima | | 分子名称: | Sugar ABC transporter, periplasmic sugar-binding protein, beta-D-glucopyranose | | 著者 | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-08-08 | | 公開日 | 2007-08-28 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of a periplasmic glucose-binding protein from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7MHH
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 277 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1908 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHN
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 277 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1908 Å) | | 主引用文献 | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MHJ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K and High Humidity | | 分子名称: | 3C-like proteinase, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.0005 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHO
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 298 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MHP
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 298 K at high humidity | | 分子名称: | 3C-like proteinase, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.0005 Å) | | 主引用文献 | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MHI
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHQ
 
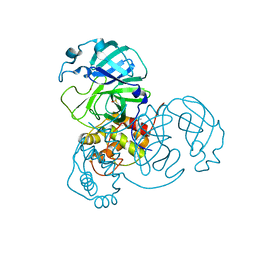 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 310 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9601 Å) | | 主引用文献 | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MRR
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Leupeptin | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, LEUPEPTIN | | 著者 | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2021-05-08 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MHK
 
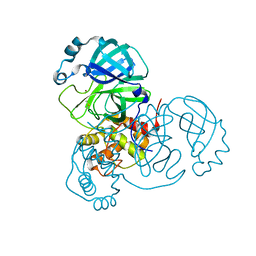 | | Crystal Structure of Apo/Unliganded SARS-CoV-2 Main Protease (Mpro) at 310 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9601 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | 分子名称: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2021-04-30 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
2QXY
 
 | |
2UW1
 
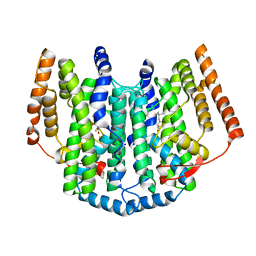 | | Ivy Desaturase Structure | | 分子名称: | (3R)-3-HYDROXY-5,5-DIMETHYLHEXANOIC ACID, FE (III) ION, PLASTID DELTA4 MULTIFUNCTIONAL ACYL-ACYL CARRIER PROTEIN DESATURASE, ... | | 著者 | Guy, J.E, Whittle, E, Kumaran, D, Lindqvist, Y, Shanklin, J. | | 登録日 | 2007-03-15 | | 公開日 | 2007-05-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The Crystal Structure of the Ivy {Delta}4-16:0-Acp Desaturase Reveals Structural Details of the Oxidized Active Site and Potential Determinants of Regioselectivity.
J.Biol.Chem., 282, 2007
|
|
1PV1
 
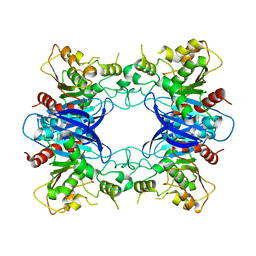 | | Crystal Structure Analysis of Yeast Hypothetical Protein: YJG8_YEAST | | 分子名称: | Hypothetical 33.9 kDa esterase in SMC3-MRPL8 intergenic region | | 著者 | Millard, C, Kumaran, D, Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-06-26 | | 公開日 | 2004-11-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural characterization and reversal of the natural organophosphate resistance of a D-type esterase, Saccharomyces cerevisiae S-formylglutathione hydrolase.
Biochemistry, 47, 2008
|
|
3NQB
 
 | |
3D3A
 
 | |
3OVG
 
 | | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound | | 分子名称: | PHOSPHATE ION, ZINC ION, amidohydrolase | | 著者 | Zhang, Z, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-09-16 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.059 Å) | | 主引用文献 | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound
To be Published
|
|
3DEB
 
 | |
3G7S
 
 | |
3GBV
 
 | | Crystal structure of a putative LacI transcriptional regulator from Bacteroides fragilis | | 分子名称: | 1,2-ETHANEDIOL, Putative LacI-family transcriptional regulator, SODIUM ION | | 著者 | Syed Ibrahim, B, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-02-20 | | 公開日 | 2009-03-10 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a putative LacI transcriptional regulator from Bacteroides fragilis
To be Published
|
|
3GRA
 
 | | Crystal structure of AraC family transcriptional regulator from Pseudomonas putida | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Bagaria, A, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-03-25 | | 公開日 | 2009-04-14 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of AraC family transcriptional regulator from Pseudomonas putida
To be Published
|
|
3GPV
 
 | | Crystal structure of a transcriptional regulator, MerR family from Bacillus thuringiensis | | 分子名称: | Transcriptional regulator, MerR family | | 著者 | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-03-23 | | 公開日 | 2009-04-14 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of a transcriptional regulator, MerR family from Bacillus thuringiensis
To be Published
|
|
3GVX
 
 | |
3SMD
 
 | |
