3QG1
 
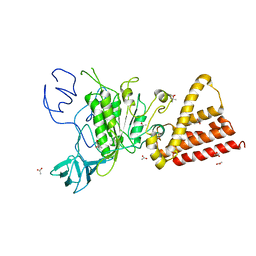 | | Crystal structure of P-loop G239A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, V-type ATP synthase alpha chain | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-24 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
8JFS
 
 | | Phosphate bound acylphosphatase from Deinococcus radiodurans at 1 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Acylphosphatase, CITRIC ACID, ... | | Authors: | Khakerwala, Z, Kumar, A, Makde, R.D. | | Deposit date: | 2023-05-18 | | Release date: | 2023-06-14 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of phosphate bound Acyl phosphatase mini-enzyme from Deinococcus radiodurans at 1 angstrom resolution.
Biochem.Biophys.Res.Commun., 671, 2023
|
|
8IYM
 
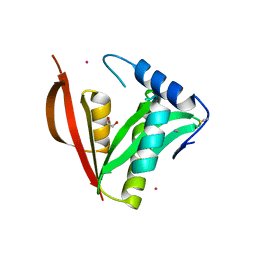 | | Crystal structure of a protein acetyltransferase, HP0935 | | Descriptor: | 1,2-ETHANEDIOL, N-acetyltransferase domain-containing protein, POTASSIUM ION, ... | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935
To be published
|
|
8IYO
 
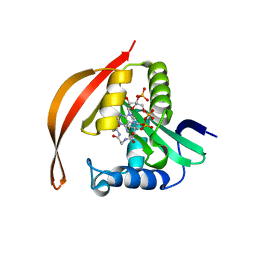 | | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase domain-containing protein | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form
To be published
|
|
4B2Z
 
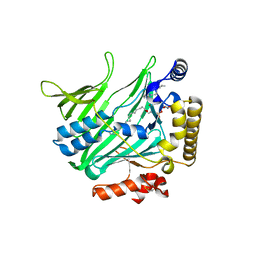 | | Structure of Osh6 in complex with phosphatidylserine | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Maeda, K, Anand, K, Chiapparino, A, Kumar, A, Poletto, M, Kaksonen, M, Gavin, A.C. | | Deposit date: | 2012-07-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interactome Map Uncovers Phosphatidylserine Transport by Oxysterol-Binding Proteins
Nature, 501, 2013
|
|
7VMI
 
 | |
7VMH
 
 | |
5GIQ
 
 | | Xaa-Pro peptidase from Deinococcus radiodurans, Zinc bound | | Descriptor: | PHOSPHATE ION, Proline dipeptidase, ZINC ION | | Authors: | Are, V.N, Singh, R, Kumar, A, Ghosh, B, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases.
Proteins, 2018
|
|
5GIV
 
 | | Crystal structure of M32 carboxypeptidase from Deinococcus radiodurans R1 | | Descriptor: | ACETATE ION, Carboxypeptidase 1, ZINC ION | | Authors: | Sharma, B, Singh, R, Yadav, P, Ghosh, B, Kumar, A, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active site gate of M32 carboxypeptidases illuminated by crystal structure and molecular dynamics simulations
Biochim. Biophys. Acta, 1865, 2017
|
|
5GIU
 
 | | Crystal structure of Xaa-Pro peptidase from Deinococcus radiodurans, metal-free active site | | Descriptor: | PHOSPHATE ION, Proline dipeptidase, SODIUM ION | | Authors: | Are, V.N, Kumar, A, Singh, R, Ghosh, B, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-25 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Xaa-Pro peptidase from Deinococcus radiodurans, metal-free active site
To Be Published
|
|
5GJL
 
 | | Solution structure of SUMO from Plasmodium falciparum | | Descriptor: | Uncharacterized protein | | Authors: | Singh, J.S, Shukla, V.K, Gujrati, M, Mishra, R.K, Kumar, A. | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interaction study of SUMO from Plasmodium falciparum
To Be Published
|
|
5I4F
 
 | | scFv 2D10 complexed with alpha 1,6 mannobiose | | Descriptor: | alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, scFv 2D10 | | Authors: | Vashisht, S, Kumar, A, Kaur, K.J, Salunke, D.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Antibodies Can Exploit Molecular Crowding to Bind New Antigens at Noncanonical Paratope Positions
CHEMISTRYSELECT, 1, 2016
|
|
6HUE
 
 | | ParkinS65N | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | McWilliams, T.G, Barini, E, Pohjolan-Pirhonen, R, Brooks, S.P, Singh, F, Burel, S, Balk, K, Kumar, A, Montava-Garriga, L, Prescott, A.R, Hassoun, S.M, Mouton-Liger, F, Ball, G, Hills, R, Knebel, A, Ulusoy, A, Di Monte, D.A, Tamjar, J, Antico, O, Fears, K, Smith, L, Brambilla, R, Palin, E, Valori, M, Eerola-Rautio, J, Tienari, P, Corti, O, Dunnett, S.B, Ganley, I.G, Suomalainen, A, Muqit, M.M.K. | | Deposit date: | 2018-10-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Phosphorylation of Parkin at serine 65 is essential for its activation in vivo .
Open Biology, 8, 2018
|
|
6IDN
 
 | | Crystal structure of ICChI chitinase from ipomoea carnea | | Descriptor: | CALCIUM ION, ICChI, a glycosylated chitinase, ... | | Authors: | Kumar, S, Kumar, A, Patel, A.K. | | Deposit date: | 2018-09-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | TIM barrel fold and glycan moieties in the structure of ICChI, a protein with chitinase and lysozyme activity.
Phytochemistry, 170, 2020
|
|
7QV7
 
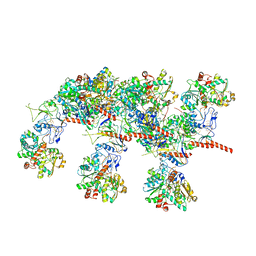 | | Cryo-EM structure of Hydrogen-dependent CO2 reductase. | | Descriptor: | Hydrogen dependent carbon dioxide reductase subunit FdhF, Hydrogen dependent carbon dioxide reductase subunit HycB3, Hydrogen dependent carbon dioxide reductase subunit HycB4, ... | | Authors: | Dietrich, H.M, Righetto, R.D, Kumar, A, Wietrzynski, W, Schuller, S.K, Trischler, R, Wagner, J, Schwarz, F.M, Engel, B.D, Mueller, V, Schuller, J.M. | | Deposit date: | 2022-01-19 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Membrane-anchored HDCR nanowires drive hydrogen-powered CO 2 fixation.
Nature, 607, 2022
|
|
6A8Z
 
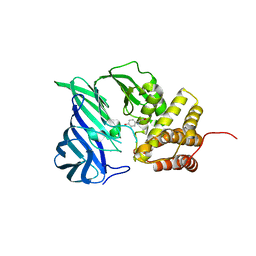 | | Crystal structure of M1 zinc metallopeptidase from Deinococcus radiodurans | | Descriptor: | SODIUM ION, TYROSINE, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Makde, R.D. | | Deposit date: | 2018-07-11 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | Two-domain aminopeptidase of M1 family: Structural features for substrate binding and gating in absence of C-terminal domain.
J.Struct.Biol., 208, 2019
|
|
4JC4
 
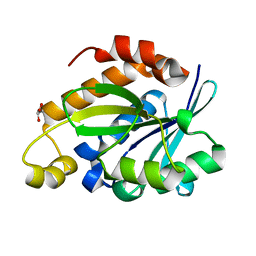 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 2.25 angstrom resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2013-02-21 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
4FNO
 
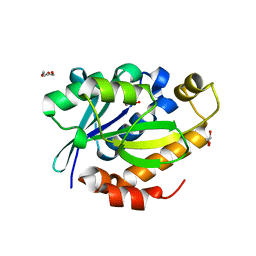 | | Crystal structure of peptidyl t-RNA hydrolase from Pseudomonas aeruginosa at 2.2 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Arora, A, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
1FMQ
 
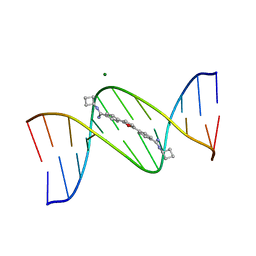 | | Cyclo-butyl-bis-furamidine complexed with dCGCGAATTCGCG | | Descriptor: | 2,5-BIS{[4-(N-CYCLOBUTYLDIAMINOMETHYL)PHENYL]}FURAN, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Simpson, I.J, Lee, M, Kumar, A, Boykin, D.W, Neidle, S. | | Deposit date: | 2000-08-18 | | Release date: | 2000-09-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA minor groove interactions and the biological activity of 2,5-bis-[4-(N-alkylamidino)phenyl] furans
Bioorg.Med.Chem.Lett., 10, 2000
|
|
1FMS
 
 | | Structure of complex between cyclohexyl-bis-furamidine and d(CGCGAATTCGCG) | | Descriptor: | 2,5-BIS{[4-(N-CYCLOHEXYLDIAMINOMETHYL)PHENYL]}FURAN, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Simpson, I.J, Lee, M, Kumar, A, Boykin, D.W, Neidle, S. | | Deposit date: | 2000-08-18 | | Release date: | 2000-09-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA minor groove interactions and the biological activity of 2,5-bis-[4-(N-alkylamidino)phenyl] furans
Bioorg.Med.Chem.Lett., 10, 2000
|
|
2BPC
 
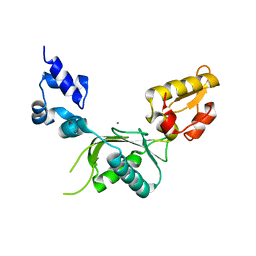 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | Descriptor: | DNA POLYMERASE BETA, MANGANESE (II) ION | | Authors: | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-07-08 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
2LGJ
 
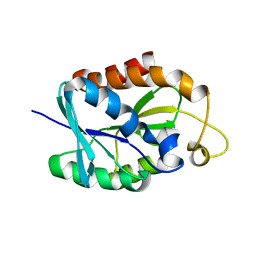 | | Solution structure of MsPTH | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Yadav, R, Pathak, P, Pulavarti, S, Jain, A, Kumar, A, Shukla, V, Arora, A. | | Deposit date: | 2011-07-27 | | Release date: | 2012-08-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of Peptidyl t-RNA hydrolase from Mycobacterium smegmatis
To be Published
|
|
6I1D
 
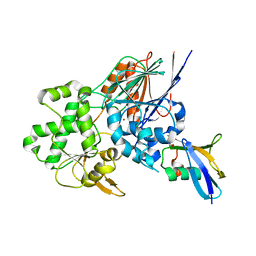 | | Structure of the Ysh1-Mpe1 nuclease complex from S.cerevisiae | | Descriptor: | Endoribonuclease YSH1, GLYCEROL, Protein MPE1, ... | | Authors: | Hill, C.H, Boreikaite, V, Kumar, A, Casanal, A, Kubik, P, Degliesposti, G, Maslen, S, Mariani, A, von Loeffelholz, O, Girbig, M, Skehel, M, Passmore, L.A. | | Deposit date: | 2018-10-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Activation of the Endonuclease that Defines mRNA 3' Ends Requires Incorporation into an 8-Subunit Core Cleavage and Polyadenylation Factor Complex.
Mol.Cell, 73, 2019
|
|
7CAY
 
 | | Crystal Structure of Lon N-terminal domain protein from Xanthomonas campestris | | Descriptor: | ATP-dependent protease | | Authors: | Singh, R, Sharma, B, Deshmukh, S, Kumar, A, Makde, R.D. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of XCC3289 from Xanthomonas campestris: homology with the N-terminal substrate-binding domain of Lon peptidase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7CLE
 
 | | Non-Specific Class-c acidphosphatase from Sphingobium sp. RSMS | | Descriptor: | Acid phosphatase, MAGNESIUM ION | | Authors: | Gaur, N.K, Kumar, A, Sunder, S, Mukhopadhyaya, R, Makde, R.D. | | Deposit date: | 2020-07-20 | | Release date: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Non-Specific Class-c acidphosphatase from Sphingobium sp. RSMS
To Be Published
|
|
