3WFZ
 
 | | Crystal structure of Galacto-N-Biose/Lacto-N-Biose I Phosphorylase C236Y Mutant | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Lacto-N-biose phosphorylase | | 著者 | Koyama, Y, Hidaka, M, Kawakami, M, Nishimoto, M, Kitaoka, M. | | 登録日 | 2013-07-25 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Directed evolution to enhance thermostability of galacto-N-biose/lacto-N-biose I phosphorylase.
Protein Eng.Des.Sel., 26, 2013
|
|
6NN6
 
 | | Structure of Dot1L-H2BK120ub nucleosome complex | | 分子名称: | DNA (145-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Anderson, C.J, Baird, M.R, Hsu, A, Barbour, E.H, Koyama, Y, Borgnia, M.J, McGinty, R.K. | | 登録日 | 2019-01-14 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural Basis for Recognition of Ubiquitylated Nucleosome by Dot1L Methyltransferase.
Cell Rep, 26, 2019
|
|
1JWQ
 
 | | Structure of the catalytic domain of CwlV, N-acetylmuramoyl-L-alanine amidase from Bacillus(Paenibacillus) polymyxa var.colistinus | | 分子名称: | N-ACETYLMURAMOYL-L-ALANINE AMIDASE CwlV, ZINC ION | | 著者 | Yamane, T, Koyama, Y, Nojiri, Y, Hikage, T, Akita, M, Suzuki, A, Shirai, T, Ise, F, Shida, T, Sekiguchi, J. | | 登録日 | 2001-09-05 | | 公開日 | 2003-11-18 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Structure of the catalytic domain of N-acetylmuramoyl-L-alanine amidase, a cell wall hydrolase from Bacillus polymyxa var.colistinus and its resemblance to the structure of carboxypeptidases
To be Published
|
|
1ZOV
 
 | | Crystal Structure of Monomeric Sarcosine Oxidase from Bacillus sp. NS-129 | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase | | 著者 | Nagata, K, Sasaki, H, Ohtsuka, J, Hua, M, Okai, M, Kubota, K, Kamo, M, Ito, K, Ichikawa, T, Koyama, Y, Tanokura, M. | | 登録日 | 2005-05-14 | | 公開日 | 2006-05-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Crystal structure of monomeric sarcosine oxidase from Bacillus sp. NS-129 reveals multiple conformations at the active-site loop
PROC.JPN.ACAD.,SER.B, 81, 2005
|
|
7TAN
 
 | | Structure of VRK1 C-terminal tail bound to nucleosome core particle | | 分子名称: | Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, Histone H3.2, ... | | 著者 | Spangler, C.J, Budziszewski, G.R, McGinty, R.K. | | 登録日 | 2021-12-21 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Multivalent DNA and nucleosome acidic patch interactions specify VRK1 mitotic localization and activity.
Nucleic Acids Res., 50, 2022
|
|
2D24
 
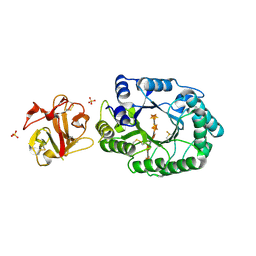 | | Crystal structure of ES complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D22
 
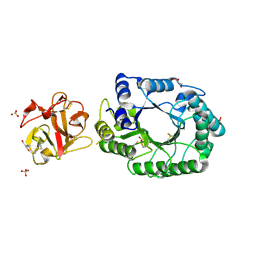 | | Crystal structure of covalent glycosyl-enzyme intermediate of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D20
 
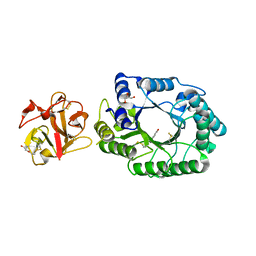 | | Crystal structure of michaelis complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, P-NITROPHENOL, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D23
 
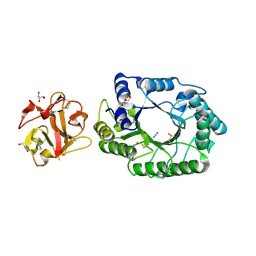 | | Crystal structure of EP complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | AZIDE ION, ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, ... | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
2D1Z
 
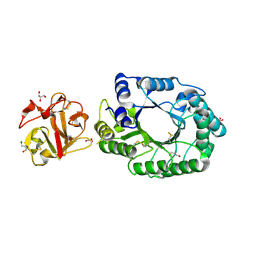 | | Crystal structure of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | 分子名称: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION | | 著者 | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | 登録日 | 2005-09-02 | | 公開日 | 2006-10-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
