3TOC
 
 | | Crystal structure of Streptococcus pyogenes Csn2 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Putative uncharacterized protein | | Authors: | Bae, E, Jung, D.K, Koo, Y. | | Deposit date: | 2011-09-05 | | Release date: | 2012-05-30 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal Structure of Streptococcus pyogenes Csn2 Reveals Calcium-Dependent Conformational Changes in Its Tertiary and Quaternary Structure
Plos One, 7, 2012
|
|
3V7F
 
 | | Crystal Structure of Streptococcus pyogenes Csn2 | | Descriptor: | CALCIUM ION, Putative uncharacterized protein | | Authors: | Bae, E, Jung, D.K, Koo, Y. | | Deposit date: | 2011-12-21 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Streptococcus pyogenes Csn2 reveals calcium-dependent conformational changes in its tertiary and quaternary structure
Plos One, 7, 2012
|
|
3VZI
 
 | | Crystal Structure of CRISPR-associated Protein | | Descriptor: | Crispr-associated protein Cas5, dvulg subtype | | Authors: | Bae, E, Koo, Y. | | Deposit date: | 2012-10-12 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Conservation and Variability in the Structure and Function of the Cas5d Endoribonuclease in the CRISPR-Mediated Microbial Immune System
J.Mol.Biol., 425, 2013
|
|
3WDK
 
 | | Crystal structure of 4-phosphopantoate-beta-alanine ligase complexed with reaction intermediate | | Descriptor: | 4-phosphopantoate--beta-alanine ligase, 5'-O-[(S)-hydroxy{[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]oxy}phosphoryl]adenosine, CITRATE ANION | | Authors: | Kishimoto, A, Kita, A, Ishibashi, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of phosphopantothenate synthetase from Thermococcus kodakarensis
Proteins, 82, 2014
|
|
3WDM
 
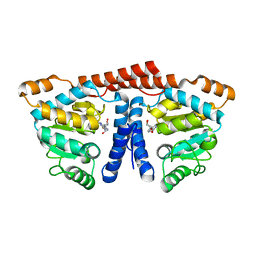 | | Crystal structure of 4-phosphopantoate-beta-alanine ligase from Thermococcus kodakarensis | | Descriptor: | 4-phosphopantoate--beta-alanine ligase, ADENOSINE | | Authors: | Kishimoto, A, Kita, A, Ishibashi, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphopantothenate synthetase from Thermococcus kodakarensis
Proteins, 82, 2014
|
|
3WDL
 
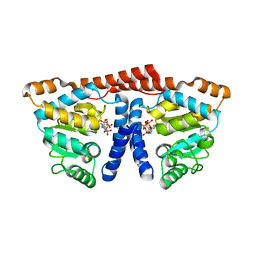 | | Crystal structure of 4-phosphopantoate-beta-alanine ligase complexed with ATP | | Descriptor: | 4-phosphopantoate--beta-alanine ligase, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Kishimoto, A, Kita, A, Ishibashi, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of phosphopantothenate synthetase from Thermococcus kodakarensis
Proteins, 82, 2014
|
|
6JBC
 
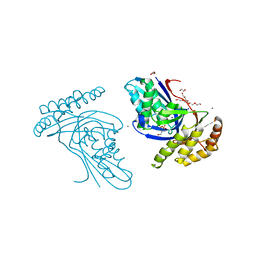 | | Phosphotransferase related to CoA biosynthesis pathway | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Kita, A, Kishimoto, A, Shimosaka, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pantoate kinase from Thermococcus kodakarensis.
Proteins, 88, 2020
|
|
6JBD
 
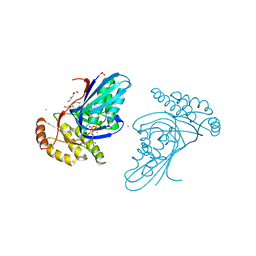 | | Phosphotransferase-ATP complex related to CoA biosynthesis pathway | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kita, A, Kishimoto, A, Shimosaka, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of pantoate kinase from Thermococcus kodakarensis.
Proteins, 88, 2020
|
|
3VZH
 
 | | Crystal Structure of CRISPR-associated Protein | | Descriptor: | Putative uncharacterized protein | | Authors: | Bae, E, Yoon, K. | | Deposit date: | 2012-10-12 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conservation and Variability in the Structure and Function of the Cas5d Endoribonuclease in the CRISPR-Mediated Microbial Immune System
J.Mol.Biol., 425, 2013
|
|
5X3R
 
 | | Crystal structure of the SmcR complexed with QStatin | | Descriptor: | 1-(5-bromanylthiophen-2-yl)sulfonylpyrazole, LuxR family transcriptional regulator, SULFATE ION | | Authors: | Jang, S.Y, Hwang, J, Kim, M.H. | | Deposit date: | 2017-02-07 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | QStatin, a Selective Inhibitor of Quorum Sensing inVibrioSpecies
MBio, 9, 2018
|
|
7OK7
 
 | | Crystal structure of the UNC119B ARL3 complex | | Descriptor: | 1,2-ETHANEDIOL, ADP-ribosylation factor-like protein 3, GLYCEROL, ... | | Authors: | Yelland, T, Ismail, S. | | Deposit date: | 2021-05-17 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The Structural and Biochemical Characterization of UNC119B Cargo Binding and Release Mechanisms.
Biochemistry, 60, 2021
|
|
7OK6
 
 | |
5HN4
 
 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and homoisocitrate | | Descriptor: | (1R,2S)-1-hydroxybutane-1,2,4-tricarboxylic acid, Homoisocitrate dehydrogenase, IMIDAZOLE, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
5HN6
 
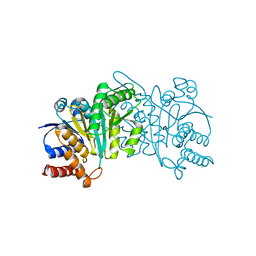 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and 3-isopropylmalate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-ISOPROPYLMALIC ACID, Homoisocitrate dehydrogenase, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
5HN5
 
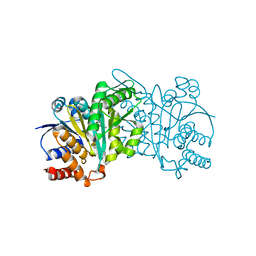 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and isocitrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Homoisocitrate dehydrogenase, ISOCITRIC ACID, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
5HN3
 
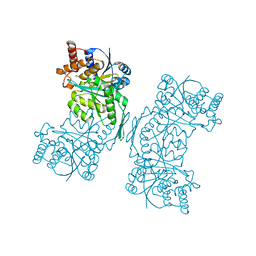 | |
