6IG8
 
 | | Crystal structure of CSF-1R kinase domain with a small molecular inhibitor, JTE-952 | | Descriptor: | (3-{4-[(4-cyclopropylphenyl)methoxy]-3-methoxyphenyl}azetidin-1-yl)(4-{[(2S)-2,3-dihydroxypropoxy]methyl}pyridin-2-yl)methanone, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Doi, S, Adachi, T. | | Deposit date: | 2018-09-25 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a novel azetidine scaffold for colony stimulating factor-1 receptor (CSF-1R) Type II inhibitors by the use of docking models.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
2ZQB
 
 | |
3WX5
 
 | |
2ZJ6
 
 | |
2ZJ7
 
 | |
3AA4
 
 | | A52V E.coli RNase HI | | Descriptor: | Ribonuclease HI | | Authors: | Takano, K. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Protein core adaptability: crystal structures of the cavity-filling variants of Escherichia coli RNase HI
Protein Pept.Lett., 17, 2010
|
|
3AIM
 
 | | R267E mutant of a HSL-like carboxylesterase from Sulfolobus tokodaii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 303aa long hypothetical esterase, ... | | Authors: | Angkawidjaja, C, Kanaya, S. | | Deposit date: | 2010-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and stability of a thermostable carboxylesterase from the thermoacidophilic archaeon Sulfolobustokodaii
Febs J., 279, 2012
|
|
3AA3
 
 | | A52L E. coli RNase HI | | Descriptor: | Ribonuclease HI | | Authors: | Takano, K. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein core adaptability: crystal structures of the cavity-filling variants of Escherichia coli RNase HI
Protein Pept.Lett., 17, 2010
|
|
3ALY
 
 | |
3AA2
 
 | | A52I E. coli RNase HI | | Descriptor: | Ribonuclease HI | | Authors: | Takano, K. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein core adaptability: crystal structures of the cavity-filling variants of Escherichia coli RNase HI
Protein Pept.Lett., 17, 2010
|
|
3AA5
 
 | | A52F E.coli RNase HI | | Descriptor: | Ribonuclease HI | | Authors: | Takano, K. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein core adaptability: crystal structures of the cavity-filling variants of Escherichia coli RNase HI
Protein Pept.Lett., 17, 2010
|
|
3AIL
 
 | |
3AIK
 
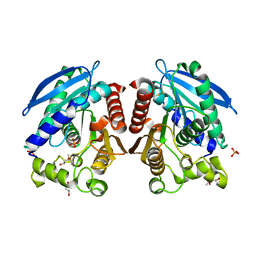 | | Crystal structure of a HSL-like carboxylesterase from Sulfolobus tokodaii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 303aa long hypothetical esterase, ... | | Authors: | Angkawidjaja, C, Kanaya, S. | | Deposit date: | 2010-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and stability of a thermostable carboxylesterase from the thermoacidophilic archaeon Sulfolobustokodaii
Febs J., 279, 2012
|
|
3AIN
 
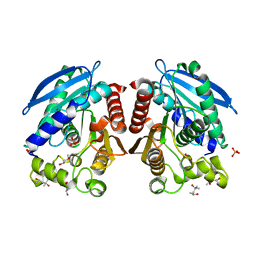 | | R267G mutant of a HSL-like carboxylesterase from Sulfolobus tokodaii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 303aa long hypothetical esterase, ... | | Authors: | Angkawidjaja, C, Kanaya, S. | | Deposit date: | 2010-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and stability of a thermostable carboxylesterase from the thermoacidophilic archaeon Sulfolobustokodaii
Febs J., 279, 2012
|
|
3AIO
 
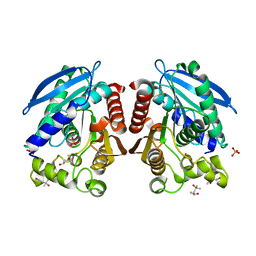 | | R267K mutant of a HSL-like carboxylesterase from Sulfolobus tokodaii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 303aa long hypothetical esterase, ... | | Authors: | Angkawidjaja, C, Kanaya, S. | | Deposit date: | 2010-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and stability of a thermostable carboxylesterase from the thermoacidophilic archaeon Sulfolobustokodaii
Febs J., 279, 2012
|
|
3ASM
 
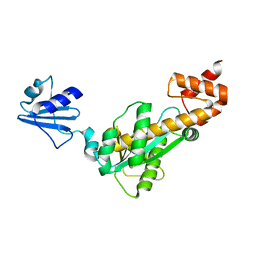 | |
