7BPP
 
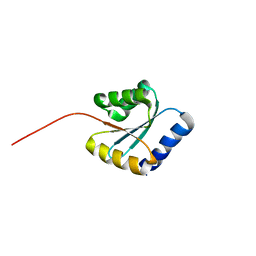 | | Solution NMR structure of NF5; de novo designed protein with a novel fold | | Descriptor: | NF5 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPL
 
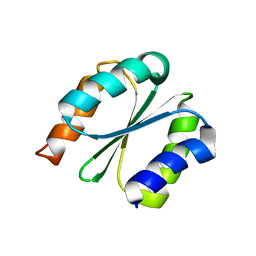 | | Solution NMR structure of NF1; de novo designed protein with a novel fold | | Descriptor: | NF1 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPN
 
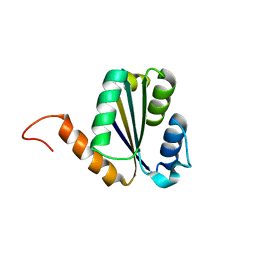 | | Solution NMR structure of NF7; de novo designed protein with a novel fold | | Descriptor: | NF7 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQE
 
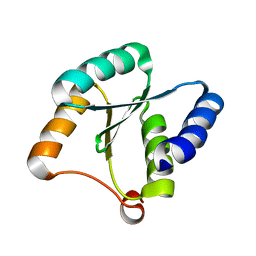 | | Solution NMR structure of NF3; de novo designed protein with a novel fold | | Descriptor: | NF3 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPM
 
 | | Solution NMR structure of NF2; de novo designed protein with a novel fold | | Descriptor: | NF2 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQD
 
 | | Solution NMR structure of NF8 (knot fold); de novo designed protein with a novel fold | | Descriptor: | NF8 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQQ
 
 | | Solution NMR structure of fold-Z Gogy; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Gogy | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-10 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQS
 
 | | Solution NMR structure of fold-U Nomur; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Nomur | | Authors: | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-10 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQR
 
 | | Solution NMR structure of fold-K Mussoc; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Mussoc | | Authors: | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-10 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQN
 
 | | Solution NMR structure of fold-C Rei; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Rei | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-10 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQC
 
 | | Solution NMR structure of NF4; de novo designed protein with a novel fold | | Descriptor: | NF4 | | Authors: | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, T, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQB
 
 | | Solution NMR structure of NF6; de novo designed protein with a novel fold | | Descriptor: | NF6 | | Authors: | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQM
 
 | | Solution NMR structure of fold-0 Chantal; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Chantal | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-10 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7DNS
 
 | | Crystal structure of domain-swapped dimer of H5_Fold-0 Elsa; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | GLYCEROL, de novo designed protein | | Authors: | Suzuki, K, Kobayashi, N, Murata, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-12-10 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
6XEH
 
 | |
7KBQ
 
 | |
5GAJ
 
 | | Solution NMR structure of De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258 | | Descriptor: | DE NOVO DESIGNED PROTEIN OR258 | | Authors: | Liu, G, Castelllanos, J, Koga, R, Koga, N, Xiao, R, Pederson, K, Janjua, H, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258
To Be Published
|
|
