7B3O
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with STE90-C11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Fab Fragment, Light Chain of Fab Fragment, ... | | Authors: | Kluenemann, T, Van den Heuvel, J. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody selected from COVID-19 patients binds to the ACE2-RBD interface and is tolerant to most known RBD mutations.
Cell Rep, 36, 2021
|
|
6I9V
 
 | |
6RTD
 
 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C, ... | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
6RTE
 
 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
6I9U
 
 | |
6I9W
 
 | | Crystal structure of the halohydrin dehalogenase HheG T123G mutant | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, Putative oxidoreductase | | Authors: | Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2018-11-26 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Position 123 of halohydrin dehalogenase HheG plays an important role in stability, activity, and enantioselectivity.
Sci Rep, 9, 2019
|
|
6TPO
 
 | | Conformation of cd1 nitrite reductase NirS without bound heme d1 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, HEME C, Nitrite reductase, ... | | Authors: | Kluenemann, T, Blankenfeldt, W. | | Deposit date: | 2019-12-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Structure of heme d 1 -free cd 1 nitrite reductase NirS.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TP9
 
 | | c-type cytochrome NirC | | Descriptor: | Cytochrome c55X, HEME C | | Authors: | Kluenemann, T, Henke, S, Blankenfeldt, W. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The crystal structure of the heme d1biosynthesis-associated small c-type cytochrome NirC reveals mixed oligomeric states in crystallo.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6TSI
 
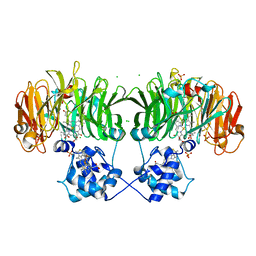 | |
6TV9
 
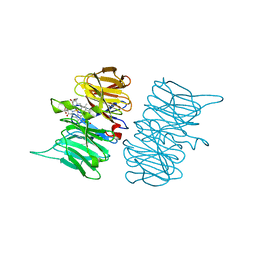 | |
6TV2
 
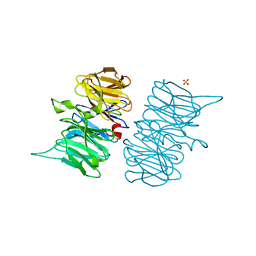 | | Heme d1 biosynthesis associated Protein NirF | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, Protein NirF, ... | | Authors: | Kluenemann, T, Layer, G, Blankenfeldt, W. | | Deposit date: | 2020-01-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Crystal structure of NirF: insights into its role in heme d 1 biosynthesis.
Febs J., 288, 2021
|
|
6Q6F
 
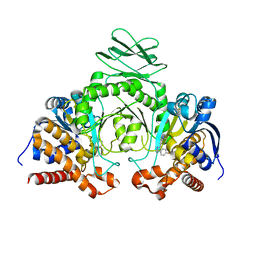 | | Crystal structure of IDH1 R132H in complex with HMS101 | | Descriptor: | (2~{R})-2-[2-[(3~{R})-3-(4-fluorophenyl)pyrrolidin-1-yl]ethyl]-1,4-dimethyl-piperazine, Isocitrate dehydrogenase [NADP] cytoplasmic | | Authors: | Chaturvedi, A, Goparaju, R, Gupta, C, Kluenemann, T, Araujo Cruz, M.M, Kloos, A, Goerlich, K, Schottmann, R, Struys, E.A, Ganser, A, Preller, M, Heuser, M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | In vivo efficacy of mutant IDH1 inhibitor HMS-101 and structural resolution of distinct binding site.
Leukemia, 34, 2020
|
|
6TUK
 
 | | Crystal structure of Fdr9 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Putative oxidoreductase, ... | | Authors: | Rodriguez, A, Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expression, purification and crystal structure determination of a ferredoxin reductase from the actinobacterium Thermobifida fusca.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TO2
 
 | | Crystal structure of CYP154C5 from Nocardia farcinica in complex with 5alpha-Androstan-3-one | | Descriptor: | (5~{S},8~{S},9~{S},10~{S},13~{S},14~{S})-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydrocyclopenta[a]phenanthren-3-one, CHLORIDE ION, Cytochrome P450 monooxygenase, ... | | Authors: | Rodriguez, A, Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CYP154C5 Regioselectivity in Steroid Hydroxylation Explored by Substrate Modifications and Protein Engineering*.
Chembiochem, 22, 2021
|
|
