1RIL
 
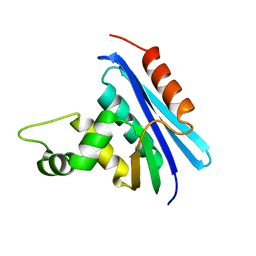 | | CRYSTAL STRUCTURE OF RIBONUCLEASE H FROM THERMUS THERMOPHILUS HB8 REFINED AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Okumura, M, Katayanagi, K, Kimura, S, Kanaya, S, Nakamura, H, Morikawa, K. | | Deposit date: | 1993-01-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ribonuclease H from Thermus thermophilus HB8 refined at 2.8 A resolution.
J.Mol.Biol., 230, 1993
|
|
2E4P
 
 | | Crystal structure of BphA3 (oxidized form) | | Descriptor: | Biphenyl dioxygenase ferredoxin subunit, FE2/S2 (INORGANIC) CLUSTER, SULFATE ION, ... | | Authors: | Senda, M, Kishigami, S, Kimura, S, Ishida, T, Fukuda, M, Senda, T. | | Deposit date: | 2006-12-15 | | Release date: | 2007-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
2E4Q
 
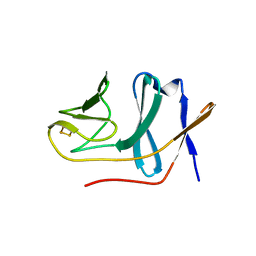 | | Crystal structure of BphA3 (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Senda, M, Kishigami, S, Kimura, S, Ishida, T, Fukuda, M, Senda, T. | | Deposit date: | 2006-12-15 | | Release date: | 2007-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
3W2E
 
 | | Crystal structure of oxidation intermediate (20 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2F
 
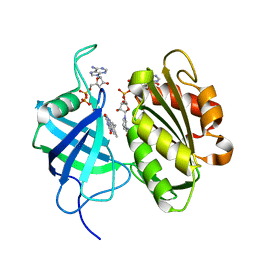 | | Crystal structure of oxidation intermediate (10 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2G
 
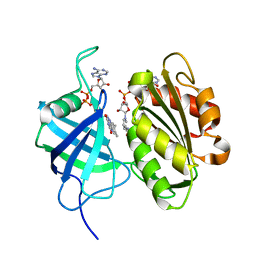 | | Crystal structure of fully reduced form of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2H
 
 | | Crystal structure of oxidation intermediate (1min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2I
 
 | | Crystal structure of re-oxidized form (60 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3X35
 
 | | Crystal structure of the reduced form of the solubilized domain of porcine cytochrome b5 in form 2 crystal | | Descriptor: | ACETATE ION, Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirano, Y, Kimura, S, Tamada, T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | High-resolution crystal structures of the solubilized domain of porcine cytochrome b5.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3X33
 
 | | Crystal structure of the oxidized form of the solubilized domain of porcine cytochrome b5 in form 2 crystal | | Descriptor: | ACETATE ION, Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirano, Y, Kimura, S, Tamada, T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | High-resolution crystal structures of the solubilized domain of porcine cytochrome b5.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3X34
 
 | | Crystal structure of the reduced form of the solubilized domain of porcine cytochrome b5 in form 1 crystal | | Descriptor: | CALCIUM ION, Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirano, Y, Kimura, S, Tamada, T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.76 Å) | | Cite: | High-resolution crystal structures of the solubilized domain of porcine cytochrome b5.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3X32
 
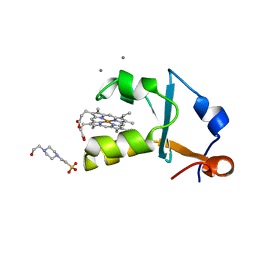 | | Crystal structure of the oxidized form of the solubilized domain of porcine cytochrome b5 in form 1 crystal | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Cytochrome b5, ... | | Authors: | Hirano, Y, Kimura, S, Tamada, T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | High-resolution crystal structures of the solubilized domain of porcine cytochrome b5.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5B86
 
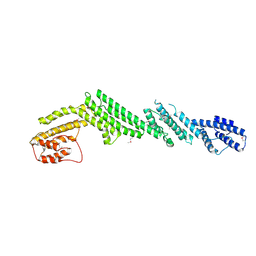 | | Crystal structure of M-Sec | | Descriptor: | Tumor necrosis factor alpha-induced protein 2 | | Authors: | Yamashita, M, Sato, Y, Yamagata, A, Fukai, S. | | Deposit date: | 2016-06-12 | | Release date: | 2016-10-12 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (3.017 Å) | | Cite: | Distinct Roles for the N- and C-terminal Regions of M-Sec in Plasma Membrane Deformation during Tunneling Nanotube Formation.
Sci Rep, 6, 2016
|
|
7P6S
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2, pentane-1,5-diol | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6T
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form III) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6R
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form I) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1LAW
 
 | |
1LAV
 
 | |
6J27
 
 | | Crystal structure of the branched-chain polyamine synthase from Thermus thermophilus (Tth-BpsA) in complex with N4-aminopropylspermidine and 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Mizohata, E, Toyoda, M, Fujita, J, Inoue, T. | | Deposit date: | 2018-12-31 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The C-terminal flexible region of branched-chain polyamine synthase facilitates substrate specificity and catalysis.
Febs J., 286, 2019
|
|
6J26
 
 | | Crystal structure of the branched-chain polyamine synthase from Thermococcus kodakarensis (Tk-BpsA) in complex with N4-bis(aminopropyl)spermidine and 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, FE (III) ION, N(4)-bis(aminopropyl)spermidine synthase, ... | | Authors: | Mizohata, E, Toyoda, M, Fujita, J, Inoue, T. | | Deposit date: | 2018-12-31 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The C-terminal flexible region of branched-chain polyamine synthase facilitates substrate specificity and catalysis.
Febs J., 286, 2019
|
|
6J28
 
 | | Crystal structure of the branched-chain polyamine synthase C9 mutein from Thermus thermophilus (Tth-BpsA C9) in complex with N4-aminopropylspermidine and 5'-methylthioadenosine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-DEOXY-5'-METHYLTHIOADENOSINE, FE (III) ION, ... | | Authors: | Mizohata, E, Toyoda, M, Fujita, J, Inoue, T. | | Deposit date: | 2018-12-31 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The C-terminal flexible region of branched-chain polyamine synthase facilitates substrate specificity and catalysis.
Febs J., 286, 2019
|
|
2GR2
 
 | |
2GQW
 
 | | Crystal structure of Ferredoxin reductase, BphA4 (oxidized form) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, GLYCEROL, ... | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2006-04-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
2GR0
 
 | |
2GR3
 
 | |
