6T9E
 
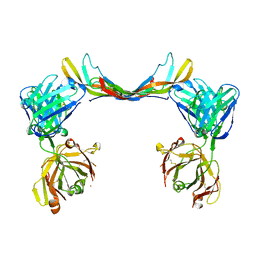 | | Crystal structure of a bispecific DutaFab in complex with human PDGF | | Descriptor: | DutaFab mat VH chain, DutaFab mat VL chain, Platelet-derived growth factor subunit B | | Authors: | Kimbung, R, Logan, D.T, Beckmann, R, Jensen, K, Speck, J, Fenn, S, Kettenberger, H. | | Deposit date: | 2019-10-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | DutaFabs are engineered therapeutic Fab fragments that can bind two targets simultaneously.
Nat Commun, 12, 2021
|
|
6T9D
 
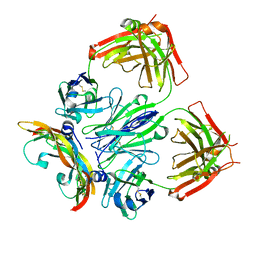 | | Crystal structure of a bispecific DutaFab in complex with human VEGF121 | | Descriptor: | VP mat DutaFab VH chain, VP mat DutaFab VL chain, Vascular endothelial growth factor A | | Authors: | Kimbung, R, Logan, D.T, Beckmann, R, Jensen, K, Speck, J, Fenn, S, Kettenberger, H. | | Deposit date: | 2019-10-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | DutaFabs are engineered therapeutic Fab fragments that can bind two targets simultaneously.
Nat Commun, 12, 2021
|
|
1EIF
 
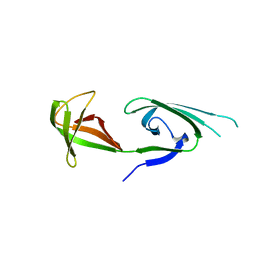 | | EUKARYOTIC TRANSLATION INITIATION FACTOR 5A FROM METHANOCOCCUS JANNASCHII | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 5A | | Authors: | Kim, K.K, Hung, L.W, Yokota, H, Kim, R, Kim, S.H. | | Deposit date: | 1998-07-29 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1LQL
 
 | | Crystal structure of OsmC like protein from Mycoplasma pneumoniae | | Descriptor: | osmotical inducible protein C like family | | Authors: | Choi, I.-G, Shin, D.H, Brandsen, J, Jancarik, J, Kim, R, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-05-10 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of a stress inducible protein from Mycoplasma pneumoniae at 2.85 A resolution
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
1Z0Z
 
 | | Crystal structure of a NAD kinase from Archaeoglobus fulgidus in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable inorganic polyphosphate/ATP-NAD kinase | | Authors: | Liu, J, Lou, Y, Yokota, H, Adams, P.D, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-03-02 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of an NAD Kinase from Archaeoglobus fulgidus in Complex with ATP, NAD, or NADP
J.Mol.Biol., 354, 2005
|
|
1L7P
 
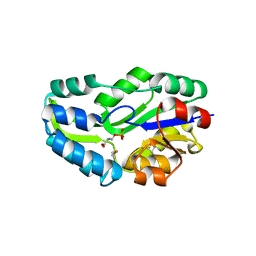 | | SUBSTRATE BOUND PHOSPHOSERINE PHOSPHATASE COMPLEX STRUCTURE | | Descriptor: | PHOSPHATE ION, PHOSPHOSERINE, PHOSPHOSERINE PHOSPHATASE | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7N
 
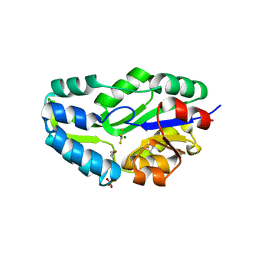 | | TRANSITION STATE ANALOGUE OF PHOSPHOSERINE PHOSPHATASE (ALUMINUM FLUORIDE COMPLEX) | | Descriptor: | ALUMINUM FLUORIDE, MAGNESIUM ION, PHOSPHOSERINE PHOSPHATASE, ... | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1Z0U
 
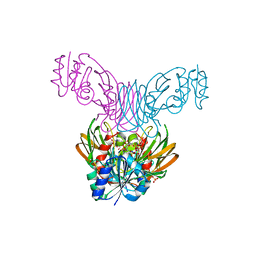 | | Crystal structure of a NAD kinase from Archaeoglobus fulgidus bound by NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable inorganic polyphosphate/ATP-NAD kinase, SULFATE ION | | Authors: | Liu, J, Lou, Y, Yokota, H, Adams, P.D, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-03-02 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of an NAD Kinase from Archaeoglobus fulgidus in Complex with ATP, NAD, or NADP
J.Mol.Biol., 354, 2005
|
|
1L2F
 
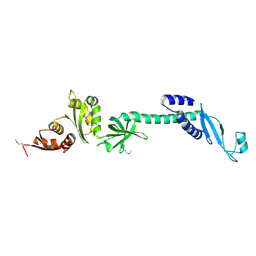 | | Crystal structure of NusA from Thermotoga maritima: a structure-based role of the N-terminal domain | | Descriptor: | N utilization substance protein A | | Authors: | Shin, D.H, Nguyen, H.H, Jancarik, J, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-02-20 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of NusA from Thermotoga maritima and functional implication of the N-terminal domain.
Biochemistry, 42, 2003
|
|
1LFP
 
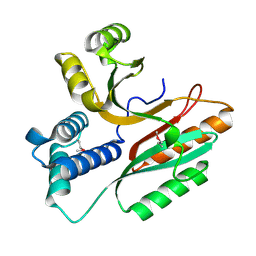 | | Crystal Structure of a Conserved Hypothetical Protein Aq1575 from Aquifex Aeolicus | | Descriptor: | Hypothetical protein AQ_1575 | | Authors: | Shin, D.H, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-04-11 | | Release date: | 2002-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of conserved hypothetical protein Aq1575 from Aquifex aeolicus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1L7O
 
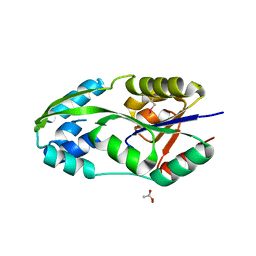 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE IN APO FORM | | Descriptor: | ACETIC ACID, PHOSPHOSERINE PHOSPHATASE, ZINC ION | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1FBN
 
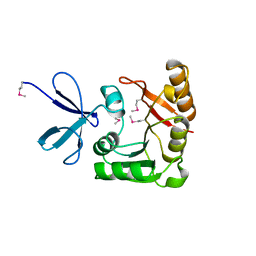 | | CRYSTAL STRUCTURE OF A FIBRILLARIN HOMOLOGUE FROM METHANOCOCCUS JANNASCHII, A HYPERTHERMOPHILE, AT 1.6 A | | Descriptor: | MJ FIBRILLARIN HOMOLOGUE | | Authors: | Wang, H, Boisvert, D, Kim, K.K, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1999-04-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution.
EMBO J., 19, 2000
|
|
1F5S
 
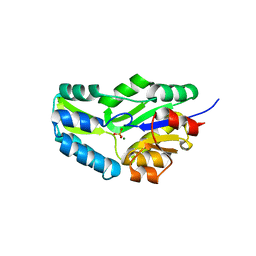 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE FROM METHANOCOCCUS JANNASCHII | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOSERINE PHOSPHATASE (PSP) | | Authors: | Wang, W, Kim, R, Jancarik, J, Yokota, H, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-06-15 | | Release date: | 2001-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 A resolution.
Structure, 9, 2001
|
|
1G2I
 
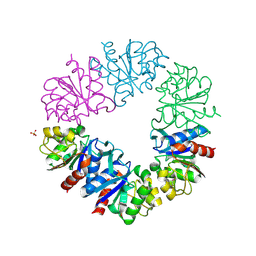 | | CRYSTAL STRUCTURE OF A NOVEL INTRACELLULAR PROTEASE FROM PYROCOCCUS HORIKOSHII AT 2 A RESOLUTION | | Descriptor: | PROTEASE I, SULFATE ION | | Authors: | Du, X, Choi, I.-G, Kim, R, Jancarik, J, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-A resolution.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2BA2
 
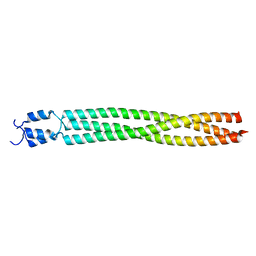 | | Crystal structure of the DUF16 domain of MPN010 from Mycoplasma pneumoniae | | Descriptor: | Hypothetical UPF0134 protein MPN010 | | Authors: | Shin, D.H, Kim, J.-S, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-10-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the DUF16 domain of MPN010 from Mycoplasma pneumoniae.
Protein Sci., 15, 2006
|
|
1ZXJ
 
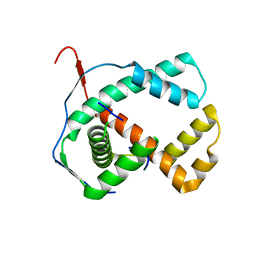 | | Crystal structure of the hypthetical Mycoplasma protein, MPN555 | | Descriptor: | Hypothetical protein MG377 homolog | | Authors: | Schulze-Gahmen, U, Aono, S, Shengfeng, C, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-06-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the hypothetical Mycoplasma protein MPN555 suggests a chaperone function.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2EIF
 
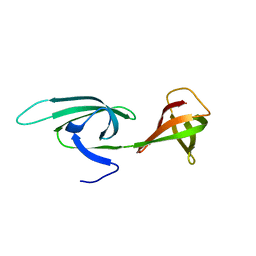 | | Eukaryotic translation initiation factor 5A from Methanococcus jannaschii | | Descriptor: | PROTEIN (EUKARYOTIC TRANSLATION INITIATION FACTOR 5A) | | Authors: | Kim, K.K, Hung, L.W, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1998-10-12 | | Release date: | 1999-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1SHS
 
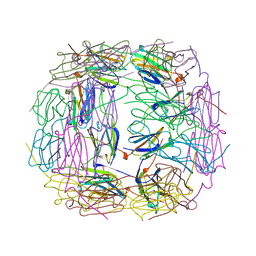 | |
1YF2
 
 | | Three-dimensional structure of DNA sequence specificity (S) subunit of a type I restriction-modification enzyme and its functional implications | | Descriptor: | Type I restriction-modification enzyme, S subunit | | Authors: | Kim, J.S, Degiovanni, A, Jancarik, J, Adams, P.D, Yokota, H.A, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-12-30 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of DNA sequence specificity subunit of a type I restriction-modification enzyme and its functional implications.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2AHD
 
 | |
1FO5
 
 | | SOLUTION STRUCTURE OF REDUCED MJ0307 | | Descriptor: | THIOREDOXIN | | Authors: | Cave, J.W, Cho, H.S, Batchelder, A.M, Kim, R, Yokota, H, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-08-24 | | Release date: | 2001-04-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution nuclear magnetic resonance structure of a protein disulfide oxidoreductase from Methanococcus jannaschii.
Protein Sci., 10, 2001
|
|
1J97
 
 | | Phospho-Aspartyl Intermediate Analogue of Phosphoserine phosphatase | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphoserine Phosphatase | | Authors: | Cho, H, Wang, W, Kim, R, Yokota, H, Damo, S, Kim, S.-H, Wemmer, D, Kustu, S, Yan, D, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2001-05-24 | | Release date: | 2001-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | BeF(3)(-) acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF(3)(-) complex with phosphoserine phosphatase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1MJH
 
 | | Structure-based assignment of the biochemical function of hypothetical protein MJ0577: A test case of structural genomics | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PROTEIN (ATP-BINDING DOMAIN OF PROTEIN MJ0577) | | Authors: | Zarembinski, T.I, Hung, L.-W, Mueller-Dieckmann, H.J, Kim, K.-K, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1998-11-04 | | Release date: | 1998-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based assignment of the biochemical function of a hypothetical protein: a test case of structural genomics.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1T6S
 
 | | Crystal structure of a conserved hypothetical protein from Chlorobium tepidum | | Descriptor: | NITRATE ION, conserved hypothetical protein | | Authors: | Kim, J.S, Shin, D.H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of ScpB from Chlorobium tepidum, a protein involved in chromosome partitioning.
Proteins, 62, 2006
|
|
1S7O
 
 | | Crystal structure of putative DNA binding protein SP_1288 from Streptococcus pygenes | | Descriptor: | Hypothetical UPF0122 protein SPy1201/SpyM3_0842/SPs1042/spyM18_1152 | | Authors: | Oganesyan, V, Pufan, R, DeGiovanni, A, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-29 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of the putative DNA-binding protein SP_1288 from Streptococcus pyogenes.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
