2ZL0
 
 | | Crystal structure of H.pylori ClpP | | 分子名称: | ATP-dependent Clp protease proteolytic subunit | | 著者 | Kim, D.Y, Kim, K.K. | | 登録日 | 2008-04-02 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The structural basis for the activation and peptide recognition of bacterial ClpP
J.Mol.Biol., 379, 2008
|
|
2ZL4
 
 | |
2ZL2
 
 | |
2ZL3
 
 | | Crystal structure of H.pylori ClpP S99A | | 分子名称: | ATP-dependent Clp protease proteolytic subunit | | 著者 | Kim, D.Y, Kim, K.K. | | 登録日 | 2008-04-02 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | The structural basis for the activation and peptide recognition of bacterial ClpP
J.Mol.Biol., 379, 2008
|
|
2ACJ
 
 | | Crystal structure of the B/Z junction containing DNA bound to Z-DNA binding proteins | | 分子名称: | 5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*GP*GP*CP*GP*CP*GP*CP*G)-3', 5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*CP*AP*TP*AP*AP*AP*CP*C)-3', Double-stranded RNA-specific adenosine deaminase | | 著者 | Ha, S.C, Lowenhaupt, K, Rich, A, Kim, Y.-G, Kim, K.K. | | 登録日 | 2005-07-19 | | 公開日 | 2005-10-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases.
Nature, 437, 2005
|
|
1UM8
 
 | | Crystal structure of helicobacter pylori ClpX | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit clpX | | 著者 | Kim, D.Y, Kim, K.K. | | 登録日 | 2003-09-25 | | 公開日 | 2003-12-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of ClpX Molecular Chaperone from Helicobacter pylori
J.Biol.Chem., 278, 2003
|
|
2BCC
 
 | | STIGMATELLIN-BOUND CYTOCHROME BC1 COMPLEX FROM CHICKEN | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | 登録日 | 1998-09-18 | | 公開日 | 1999-08-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
1L1J
 
 | | Crystal structure of the protease domain of an ATP-independent heat shock protease HtrA | | 分子名称: | heat shock protease HtrA | | 著者 | Kim, D.Y, Kim, D.R, Ha, S.C, Lokanath, N.K, Hwang, H.Y, Kim, K.K. | | 登録日 | 2002-02-18 | | 公開日 | 2003-04-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of the Protease Domain of a Heat-shock Protein HtrA from Thermotoga maritima
J.BIOL.CHEM., 278, 2003
|
|
4YGQ
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | 分子名称: | Hydrolase, TERTIARY-BUTYL ALCOHOL | | 著者 | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | 登録日 | 2015-02-26 | | 公開日 | 2015-04-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YGR
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hydrolase, MAGNESIUM ION | | 著者 | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | 登録日 | 2015-02-26 | | 公開日 | 2015-04-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.703 Å) | | 主引用文献 | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YGS
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | 分子名称: | CITRIC ACID, Hydrolase, MAGNESIUM ION | | 著者 | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | 登録日 | 2015-02-26 | | 公開日 | 2015-04-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
5C5Z
 
 | | Crystal structure analysis of c4763, a uropathogenic E. coli-specific protein | | 分子名称: | Glutamyl-tRNA amidotransferase | | 著者 | Kim, H, Choi, J, Kim, D, Kim, K.K. | | 登録日 | 2015-06-22 | | 公開日 | 2015-08-19 | | 最終更新日 | 2017-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structure analysis of c4763, a uropathogenic Escherichia coli-specific protein.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1SFU
 
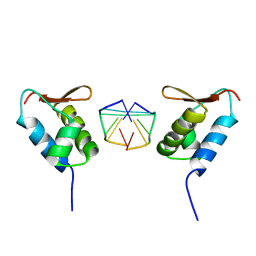 | | Crystal structure of the viral Zalpha domain bound to left-handed Z-DNA | | 分子名称: | 34L protein, 5'-D(*T*CP*GP*CP*GP*CP*G)-3' | | 著者 | Ha, S.C, Van Quyen, D, Wu, C.A, Lowenhaupt, K, Rich, A, Kim, Y.G, Kim, K.K. | | 登録日 | 2004-02-20 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1MZL
 
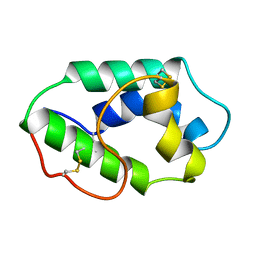 | | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | 分子名称: | MAIZE NONSPECIFIC LIPID TRANSFER PROTEIN | | 著者 | Shin, D.H, Lee, J.Y, Hwang, K.Y, Kim, K.K, Suh, S.W. | | 登録日 | 1995-01-26 | | 公開日 | 1996-08-01 | | 最終更新日 | 2018-03-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-resolution crystal structure of the non-specific lipid-transfer protein from maize seedlings.
Structure, 3, 1995
|
|
5IS1
 
 | |
5J6X
 
 | |
5JNM
 
 | | Crystal structure of MtlD from Staphylococcus aureus at 1.7-Angstrom resolution | | 分子名称: | Mannitol-1-phosphate 5-dehydrogenase, SULFATE ION | | 著者 | Ta, H.M, Nguyen, T, Kim, T, Kim, K.K. | | 登録日 | 2016-04-30 | | 公開日 | 2017-11-08 | | 最終更新日 | 2019-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Targeting Mannitol Metabolism as an Alternative Antimicrobial Strategy Based on the Structure-Function Study of Mannitol-1-Phosphate Dehydrogenase in Staphylococcus aureus.
Mbio, 10, 2019
|
|
2P52
 
 | |
4NJR
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | 分子名称: | CARBONATE ION, Probable M18 family aminopeptidase 2, ZINC ION | | 著者 | Nguyen, D.D, Pandian, R, Kim, D.Y, Ha, S.C, Yun, K.H, Kim, K.S, Kim, J.H, Kim, K.K. | | 登録日 | 2013-11-11 | | 公開日 | 2014-04-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
2P4B
 
 | | Crystal structure of E.coli RseB | | 分子名称: | Sigma-E factor regulatory protein rseB, octyl beta-D-glucopyranoside | | 著者 | Kim, D.Y, Kim, K.K. | | 登録日 | 2007-03-12 | | 公開日 | 2007-05-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of RseB and a model of its binding mode to RseA
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4NJQ
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CARBONATE ION, COBALT (II) ION, ... | | 著者 | Nguyen, D.D, Pandian, R, Kim, D.Y, Ha, S.C, Yun, K.H, Kim, K.S, Kim, J.H, Kim, K.K. | | 登録日 | 2013-11-11 | | 公開日 | 2014-04-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.702 Å) | | 主引用文献 | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4OID
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | 分子名称: | Probable M18 family aminopeptidase 2 | | 著者 | Nguyen, D.D, Pandian, R, Kim, D.D, Ha, S.C, Yoon, H.J, Kim, K.S, Yun, K.H, Kim, J.H, Kim, K.K. | | 登録日 | 2014-01-19 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
4OIW
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | 分子名称: | Probable M18 family aminopeptidase 2, ZINC ION | | 著者 | Nguyen, D.D, Pandian, R, Kim, D.D, Ha, S.C, Yoon, H.J, Kim, K.S, Yun, K.H, Kim, J.H, Kim, K.K. | | 登録日 | 2014-01-20 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
3JUJ
 
 | |
3JUK
 
 | |
