6K1G
 
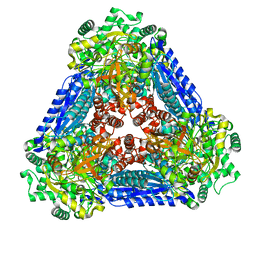 | | Crystal structure of the L-fucose isomerase soaked with Mn2+ from Raoultella sp. | | Descriptor: | L-fucose isomerase, MANGANESE (II) ION | | Authors: | Kim, I.J, Kim, D.H, Nam, K.H, Kim, K.H. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Enzymatic synthesis of l-fucose from l-fuculose using a fucose isomerase fromRaoultellasp. and the biochemical and structural analyses of the enzyme.
Biotechnol Biofuels, 12, 2019
|
|
6K1F
 
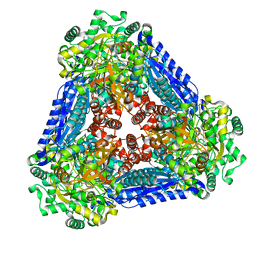 | | Crystal structure of the L-fucose isomerase from Raoultella sp. | | Descriptor: | L-fucose isomerase, MANGANESE (II) ION | | Authors: | Kim, I.J, Kim, D.H, Nam, K.H, Kim, K.H. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic synthesis of l-fucose from l-fuculose using a fucose isomerase fromRaoultellasp. and the biochemical and structural analyses of the enzyme.
Biotechnol Biofuels, 12, 2019
|
|
3V9A
 
 | |
5Z6V
 
 | |
5Y8Q
 
 | | ZsYellow at pH 8.0 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y4J
 
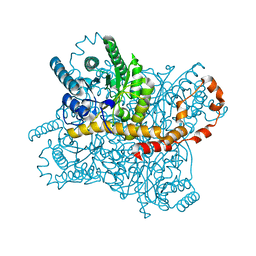 | |
5Y4I
 
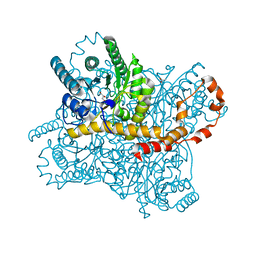 | | Crystal structure of glucose isomerase in complex with glycerol in one metal binding mode | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of glucose isomerase in complex with xylitol inhibitor in one metal binding mode
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y8R
 
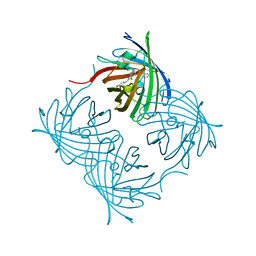 | | ZsYellow at pH 3.5 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
8IH0
 
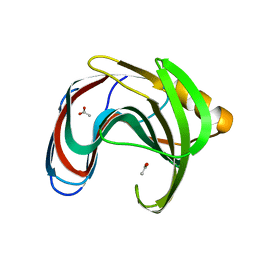 | | Crystal structure of GH11 from Thermoanaerobacterium saccharolyticum | | Descriptor: | ACETATE ION, Endo-1,4-beta-xylanase | | Authors: | Nam, K.H. | | Deposit date: | 2023-02-22 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization and structural analysis of the endo-1,4-beta-xylanase GH11 from the hemicellulose-degrading Thermoanaerobacterium saccharolyticum useful for lignocellulose saccharification.
Sci Rep, 13, 2023
|
|
8IH1
 
 | |
6LOF
 
 | | Crystal structure of ZsYellow soaked by Cu2+ | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Nam, K.H. | | Deposit date: | 2020-01-05 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Spectroscopic and Structural Analysis of Cu 2+ -Induced Fluorescence Quenching of ZsYellow.
Biosensors (Basel), 10, 2020
|
|
5HZT
 
 | | Crystal structure of Dronpa-Cu2+ | | Descriptor: | COPPER (II) ION, Fluorescent protein Dronpa | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
5HZU
 
 | | Crystal structure of Dronpa-Ni2+ | | Descriptor: | Fluorescent protein Dronpa, NICKEL (II) ION | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
5HZS
 
 | | Crystal structure of Dronpa-Co2+ | | Descriptor: | COBALT (II) ION, Fluorescent protein Dronpa | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-02-03 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
7DIG
 
 | |
7E5J
 
 | | Crystal structure of beta-glucosidase from Thermoanaerobacterium saccharolyticum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, SODIUM ION | | Authors: | Nam, K.H. | | Deposit date: | 2021-02-18 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Biochemical and Structural Analysis of a Glucose-Tolerant beta-Glucosidase from the Hemicellulose-Degrading Thermoanaerobacterium saccharolyticum.
Molecules, 27, 2022
|
|
