3FU5
 
 | |
3FUJ
 
 | |
3FTV
 
 | |
3FUM
 
 | |
3FTU
 
 | | Leukotriene A4 hydrolase in complex with dihydroresveratrol | | Descriptor: | 5-[2-(4-hydroxyphenyl)ethyl]benzene-1,3-diol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Davies, D.R. | | Deposit date: | 2009-01-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of leukotriene A4 hydrolase inhibitors using metabolomics biased fragment crystallography.
J.Med.Chem., 52, 2009
|
|
3FTS
 
 | | Leukotriene A4 hydrolase in complex with resveratrol | | Descriptor: | ACETATE ION, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Davies, D.R. | | Deposit date: | 2009-01-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of leukotriene A4 hydrolase inhibitors using metabolomics biased fragment crystallography.
J.Med.Chem., 52, 2009
|
|
3FU6
 
 | |
3FUN
 
 | |
3FTW
 
 | |
3FUD
 
 | |
3FUE
 
 | |
2LYR
 
 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYK
 
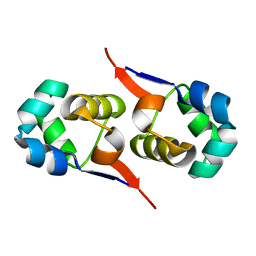 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYL
 
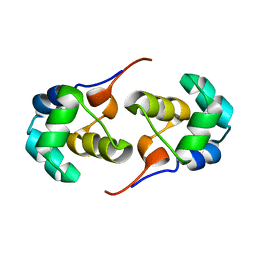 | | NOE-based 3D structure of the predissociated homodimer of CylR2 in equilibrium with monomer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYS
 
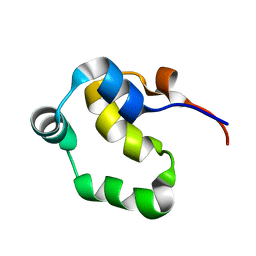 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 257K (-16 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYJ
 
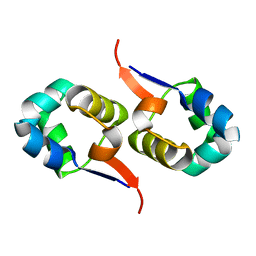 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
4FJO
 
 | | Structure of the Rev1 CTD-Rev3/7-Pol kappa RIR complex | | Descriptor: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | Wojtaszek, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2012-06-11 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol zeta and Pol kappa
J.Biol.Chem., 287, 2012
|
|
2LYP
 
 | | NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYQ
 
 | | NOE-based 3D structure of the monomeric intermediate of CylR2 at 262K (-11 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
3GW8
 
 | |
3GP5
 
 | |
3GP3
 
 | |
2K21
 
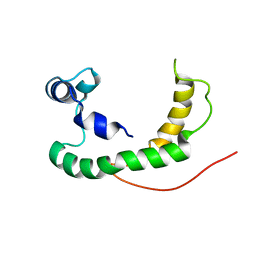 | | NMR structure of human KCNE1 in LMPG micelles at pH 6.0 and 40 degree C | | Descriptor: | Potassium voltage-gated channel subfamily E member | | Authors: | Kang, C, Tian, C, Sonnichsen, F.D, Smith, J.A, Meiler, J, George, A.L, Vanoye, C.G, Sanders, C.R, Kim, H. | | Deposit date: | 2008-03-19 | | Release date: | 2008-12-09 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel.
Biochemistry, 47, 2008
|
|
2KDC
 
 | | NMR Solution Structure of E. coli diacylglycerol kinase (DAGK) in DPC micelles | | Descriptor: | Diacylglycerol kinase | | Authors: | Van Horn, W.D, Kim, H, Ellis, C.D, Hadziselimovic, A, Sulistijo, E.S, Karra, M.D, Tian, C, Sonnichsen, F.D, Sanders, C.R. | | Deposit date: | 2009-01-06 | | Release date: | 2009-07-07 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase
Science, 324, 2009
|
|
3LNT
 
 | |
