2A7Y
 
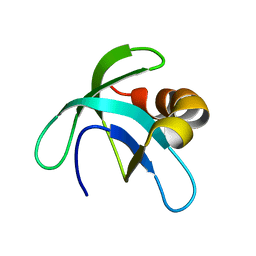 | | Solution Structure of the Conserved Hypothetical Protein Rv2302 from the Bacterium Mycobacterium tuberculosis | | Descriptor: | Hypothetical protein Rv2302/MT2359 | | Authors: | Buchko, G.W, Kim, C.-Y, Terwilliger, T.C, Kennedy, M.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-06 | | Release date: | 2005-08-23 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the conserved hypothetical protein Rv2302 from Mycobacterium tuberculosis.
J.Bacteriol., 188, 2006
|
|
2B3W
 
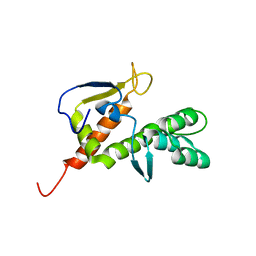 | | NMR structure of the E.coli protein YbiA, Northeast Structural Genomics target ET24. | | Descriptor: | Hypothetical protein ybiA | | Authors: | Ramelot, T.A, Cort, J.R, Xiao, R, Shih, L.Y, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-21 | | Release date: | 2005-11-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the E.coli protein YbiA, Northeast Structural Genomics target ET24.
To be Published
|
|
2AZH
 
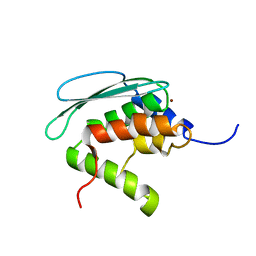 | | Solution structure of iron-sulfur cluster assembly protein SUFU from Bacillus subtilis, with zinc bound at the active site. Northeast Structural Genomics Consortium target SR17 | | Descriptor: | SufU, ZINC ION | | Authors: | Kornhaber, G.J, Swapna, G.V.T, Ramelot, T.A, Cort, J.R, Aramini, J.M, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-10 | | Release date: | 2005-09-20 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Zn-Ligated Fe-S Cluster Assembly Scaffold Protein SufU From Bacillus subtilis
To be published
|
|
2DSM
 
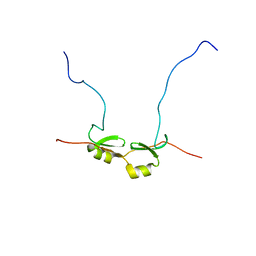 | | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450 | | Descriptor: | Hypothetical protein yqaI | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-01 | | Release date: | 2006-08-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqaI, Northeast Structural Genomics Target SR450
to be published
|
|
1JJG
 
 | | Solution Structure of Myxoma Virus Protein M156R | | Descriptor: | M156R | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-07-05 | | Release date: | 2002-03-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2alpha.
J.Mol.Biol., 322, 2002
|
|
6UT9
 
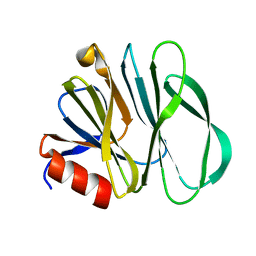 | | Crystal structure of the carbohydrate-binding domain VP8* of human P[4] rotavirus strain BM5265 | | Descriptor: | Outer capsid protein VP4 | | Authors: | Xu, S, Stuckert, M, Burnside, R, McGinnis, K, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6VKX
 
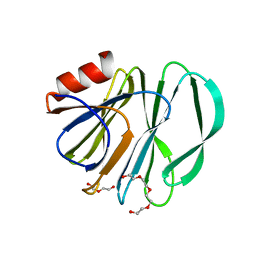 | | Crystal structure of the carbohydrate-binding domain VP8* of human P[8] rotavirus strain BM13851 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Outer capsid protein VP4, TETRAETHYLENE GLYCOL | | Authors: | Xu, S, McGinnis, K.R, Jiang, X, Kennedy, M.A. | | Deposit date: | 2020-01-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6NIW
 
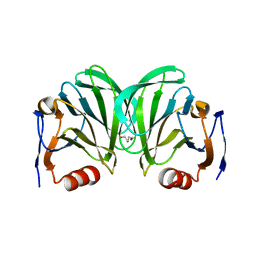 | | Crystal structure of P[6] rotavirus | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protease-sensitive outer capsid protein | | Authors: | Xu, S, Liu, Y, Lakamp, L, Ahmed, L, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens.
Plos Pathog., 16, 2020
|
|
6OAI
 
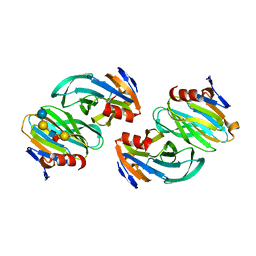 | | Crystal structure of P[6] rotavirus vp8* complexed with LNFPI | | Descriptor: | Protease-sensitive outer capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Xu, S, Liu, Y, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens.
Plos Pathog., 16, 2020
|
|
8E12
 
 | |
8E0M
 
 | |
8E0N
 
 | |
8E0O
 
 | |
8FLX
 
 | |
1D1R
 
 | |
1COC
 
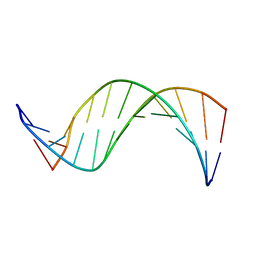 | | SOLUTION-STATE STRUCTURE OF A DNA DODECAMER DUPLEX CONTAINING A CIS-SYN THYMINE CYCLOBUTANE DIMER. | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*AP*TP*TP*CP*GP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*CP*GP*AP*AP*TP*TP*AP*AP*G)-3') | | Authors: | McAteer, K, Jing, Y, Kao, J, Taylor, J.-S, Kennedy, M.A. | | Deposit date: | 1999-05-26 | | Release date: | 1999-06-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution-state structure of a DNA dodecamer duplex containing a Cis-syn thymine cyclobutane dimer, the major UV photoproduct of DNA.
J.Mol.Biol., 282, 1998
|
|
1D4U
 
 | | INTERACTIONS OF HUMAN NUCLEOTIDE EXCISION REPAIR PROTEIN XPA WITH RPA70 AND DNA: CHEMICAL SHIFT MAPPING AND 15N NMR RELAXATION STUDIES | | Descriptor: | NUCLEOTIDE EXCISION REPAIR PROTEIN XPA (XPA-MBD), ZINC ION | | Authors: | Buchko, G.W, Daughdrill, G.W, de Lorimier, R, Rao, S, Isern, N.G, Lingbeck, J, Taylor, J, Wold, M.S, Gochin, M, Spicer, L.D, Lowry, D.F, Kennedy, M.A. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: chemical shift mapping and 15N NMR relaxation studies.
Biochemistry, 38, 1999
|
|
1EO1
 
 | |
1EIW
 
 | |
1EZE
 
 | | STRUCTURAL STUDIES OF A BABOON (PAPIO SP.) PLASMA PROTEIN INHIBITOR OF CHOLESTERYL ESTER TRANSFERASE. | | Descriptor: | CHOLESTERYL ESTER TRANSFERASE INHIBITOR PROTEIN | | Authors: | Buchko, G.W, Rozek, A, Kanda, P, Kennedy, M.A, Cushley, R.J. | | Deposit date: | 2000-05-10 | | Release date: | 2000-09-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural studies of a baboon (Papio sp.) plasma protein inhibitor of cholesteryl ester transferase.
Protein Sci., 9, 2000
|
|
1Q27
 
 | |
1RZW
 
 | | The Solution Structure of the Archaeglobus fulgidis protein AF2095. Northeast Structural Genomics Consortium target GR4 | | Descriptor: | Protein AF2095(GR4) | | Authors: | Powers, R, Acton, T.B, Huang, Y.J, Liu, J, Ma, L, Rost, B, Chiang, Y, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-29 | | Release date: | 2004-11-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes
Protein Sci., 14, 2005
|
|
1NYN
 
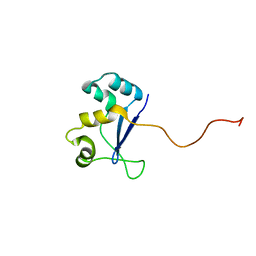 | | Solution NMR Structure of Protein YHR087W from Saccharomyces cerevisiae. Northeast Structural Genomics Consortium Target YTYST425. | | Descriptor: | Hypothetical 12.0 kDa protein in NAM8-GAR1 intergenic region | | Authors: | Cort, J.R, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-13 | | Release date: | 2003-04-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
1SOU
 
 | | NMR structure of Aquifex aeolicus 5,10-methenyltetrahydrofolate synthetase: Northeast Structural Genomics Consortium Target QR46 | | Descriptor: | 5,10-methenyltetrahydrofolate synthetase | | Authors: | Cort, J.R, Chiang, Y, Acton, T, Wu, M, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-15 | | Release date: | 2004-06-22 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Aquifex aeolicus 5,10-methenyltetrahydrofolate synthetase: Northeast Structural Genomics Consortium Target QR46
To be Published
|
|
1SSV
 
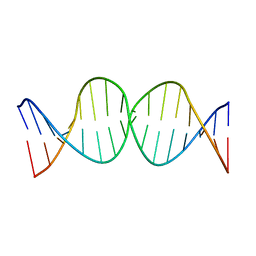 | | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | | Descriptor: | 5'-D(*CP*GP*AP*GP*GP*TP*TP*TP*AP*AP*AP*CP*CP*TP*CP*G)-3' | | Authors: | McAteer, K, Aceves-Gaona, A, Michalczyk, R, Buchko, G.W, Isern, N.G, Silks, L.A, Miller, J.H, Kennedy, M.A. | | Deposit date: | 2004-03-24 | | Release date: | 2004-12-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
Biopolymers, 75, 2004
|
|
