9NSM
 
 | | Crystal structure of the bat rotavirus apo P[10] VP8* receptor binding domain at 1.85 angstrom resolution | | Descriptor: | GLYCEROL, Outer capsid protein VP8*, SODIUM ION | | Authors: | Kennedy, M.A, Ni, S, Li, F, Wang, D, Soni, S. | | Deposit date: | 2025-03-17 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the bat rotavirus apo P[10] VP8* receptor binding domain at 1.85 angstrom resolution
To Be Published
|
|
8VX7
 
 | |
8TM9
 
 | | Computationally designed tunable C2 symmetric tandem repeat homodimer, D_3_633_8x bound to peptide | | Descriptor: | D_3_633_8x peptide bound, MC1, N-PROPANOL, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Hicks, D.R. | | Deposit date: | 2023-07-28 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computationally designed tunable C2 symmetric tandem repeat homodimer, D_3_633_8x bound to peptide
To Be Published
|
|
8TLP
 
 | | Computationally designed tunable C2 symmetric tandem repeat homodimer, D_3_633_8x without peptide | | Descriptor: | D_3_633_8x, no peptide, SULFATE ION | | Authors: | Kennedy, M.A, Stoddard, B.L, Hicks, D.R. | | Deposit date: | 2023-07-27 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computationally designed tunable C2 symmetric tandem repeat homodimer, D_3_633_8x, no peptide
To Be Published
|
|
8E0L
 
 | |
8EM1
 
 | | Type IIS Restriction Endonuclease PaqCI, DNA Unbound | | Descriptor: | 1,2-ETHANEDIOL, PaqCI, DNA Unbound | | Authors: | Kennedy, M.A, Stoddard, B.L. | | Deposit date: | 2022-09-26 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures, activity and mechanism of the Type IIS restriction endonuclease PaqCI.
Nucleic Acids Res., 51, 2023
|
|
8EPX
 
 | | Type IIS Restriction Endonuclease PaqCI, DNA bound | | Descriptor: | CALCIUM ION, DNA 1a, DNA 1b, ... | | Authors: | Kennedy, M.A, Stoddard, B.L. | | Deposit date: | 2022-10-06 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structures, activity and mechanism of the Type IIS restriction endonuclease PaqCI.
Nucleic Acids Res., 51, 2023
|
|
6OMX
 
 | |
7RKC
 
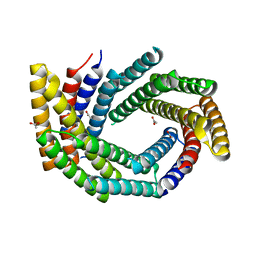 | | Computationally designed tunable C2 symmetric tandem repeat homodimer, D_3_633 | | Descriptor: | ACETATE ION, D_3_633 | | Authors: | Kennedy, M.A, Stoddard, B.L, Hicks, D.R, Bera, A.K. | | Deposit date: | 2021-07-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2KBN
 
 | | Solution NMR structure of the OB domain (residues 67-166) of MM0293 from Methanosarcina mazei. Northeast Structural Genomics Consortium target MaR214a. | | Descriptor: | Conserved protein | | Authors: | Ramelot, T.A, Ding, K, Magliqui, M, Jiang, M, Ciccosanti, C, Xiao, R, Lui, J, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-03 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the OB domain of mm0293 from the archea methanosarcina mazei
To be Published
|
|
6UV7
 
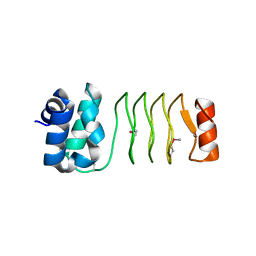 | |
6UVI
 
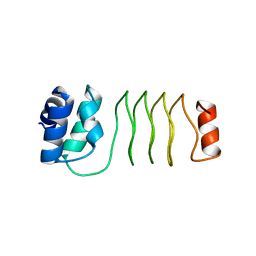 | |
7UNJ
 
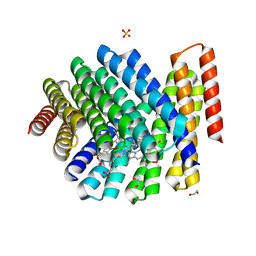 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, SP1-ZnPPaM designed chlorophyll dimer protein, SULFATE ION, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
7UNH
 
 | |
7UNI
 
 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester, SP2-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SP2-ZnPPaM designed chlorophyll dimer protein, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
3DU1
 
 | |
3E56
 
 | |
2G0Y
 
 | | Crystal Structure of a Lumenal Pentapeptide Repeat Protein from Cyanothece sp 51142 at 2.3 Angstrom Resolution. Tetragonal Crystal Form | | Descriptor: | CALCIUM ION, pentapeptide repeat protein | | Authors: | Kennedy, M.A, Ni, S, Buchko, G.W, Robinson, H. | | Deposit date: | 2006-02-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of two potentially universal turn motifs that shape the repeated five-residues fold - Crystal structure of a lumenal pentapeptide repeat protein from Cyanothece 51142
Protein Sci., 15, 2006
|
|
2F3L
 
 | |
2O5F
 
 | | Crystal Structure of DR0079 from Deinococcus radiodurans at 1.9 Angstrom Resolution | | Descriptor: | Putative Nudix hydrolase DR_0079 | | Authors: | Kennedy, M.A, Buchko, G.W, Ni, S, Robinson, H. | | Deposit date: | 2006-12-05 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and Structural Characterization of DR_0079 from Deinococcus radiodurans, a Novel Nudix Hydrolase with a Preference for Cytosine (Deoxy)ribonucleoside 5'-Di- and Triphosphates.
Biochemistry, 47, 2008
|
|
2O6W
 
 | | Crystal Structure of a Pentapeptide Repeat Protein (Rfr23) from the cyanobacterium Cyanothece 51142 | | Descriptor: | ARSENIC, Repeat Five Residue (Rfr) protein or pentapeptide repeat protein | | Authors: | Kennedy, M.A, Buchko, G.W, Ni, S, Robinson, H, Pakrasi, H.B. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the structural variation between pentapeptide repeat proteins-Crystal structure of Rfr23 from Cyanothece 51142.
J.Struct.Biol., 162, 2008
|
|
2KCM
 
 | | Solution NMR structure of the N-terminal OB-domain of SO_1732 from Shewanella oneidensis. Northeast Structural Genomics Consortium Target SoR210A. | | Descriptor: | Cold shock domain family protein | | Authors: | Ramelot, T.A, Maglaqui, M, Jiang, M, Ciccosanti, C, Xiao, R, Lui, J, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-23 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the N-terminal OB-domain of SO_1732 from Shewanella oneidensis. Northeast Structural Genomics Consortium Target SoR210A.
To be Published
|
|
2KEN
 
 | | Solution NMR structure of the OB domain (residues 67-166) of MM0293 from Methanosarcina mazei. Northeast Structural Genomics Consortium target MaR214a. | | Descriptor: | Conserved protein | | Authors: | Ramelot, T.A, Ding, K, Magliqui, M, Jiang, M, Ciccosanti, C, Xiao, R, Lui, J, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the OB domain of MM0293 from the archea Methanosarcina mazei
To be Published
|
|
1Q27
 
 | |
1YX3
 
 | | NMR structure of Allochromatium vinosum DsrC: Northeast Structural Genomics Consortium target OP4 | | Descriptor: | hypothetical protein DsrC | | Authors: | Cort, J.R, Dahl, C, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-19 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Allochromatium vinosum DsrC: solution-state NMR structure, redox properties, and interaction with DsrEFH, a protein essential for purple sulfur bacterial sulfur oxidation.
J.Mol.Biol., 382, 2008
|
|
