3NDJ
 
 | | X-ray Structure of a C-3'-Methyltransferase in Complex with S-Adenosyl-L-Homocysteine and Sugar Product | | Descriptor: | (2R,4S,6R)-4-amino-4,6-dimethyl-5-oxotetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), Methyltransferase, PHOSPHATE ION, ... | | Authors: | Bruender, N.A, Thoden, J.B, Kaur, M, Avey, M.K, Holden, H.M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Architecture of a C-3'-Methyltransferase Involved in the Biosynthesis of d-Tetronitrose.
Biochemistry, 49, 2010
|
|
3NDI
 
 | | X-ray Structure of a C-3'-Methyltransferase in Complex with S-adenosylmethionine and dTMP | | Descriptor: | Methyltransferase, PHOSPHATE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Bruender, N.A, Thoden, J.B, Kaur, M, Avey, M.K, Holden, H.M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Architecture of a C-3'-Methyltransferase Involved in the Biosynthesis of d-Tetronitrose.
Biochemistry, 49, 2010
|
|
7WBN
 
 | | PDB structure of RevCC | | Descriptor: | RevCC | | Authors: | Han, S, Kim, D, Kaur, M, Lim, Y.B, Barnwal, R.P. | | Deposit date: | 2021-12-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pseudo-Isolated alpha-Helix Platform for the Recognition of Deep and Narrow Targets.
J.Am.Chem.Soc., 144, 2022
|
|
7SEH
 
 | | Glucose-6-phosphate 1-dehydrogenase (K403QdLtL) | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mathews, I.I, Garcia, A.A, Wakatsuki, S, Mochly-Rosen, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Stabilization of glucose-6-phosphate dehydrogenase oligomers enhances catalytic activity and stability of clinical variants.
J.Biol.Chem., 298, 2022
|
|
7SEI
 
 | | Glucose-6-phosphate 1-dehydrogenase (K403Q) | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mathews, I.I, Garcia, A.A, Wakatsuki, S, Mochly-Rosen, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Stabilization of glucose-6-phosphate dehydrogenase oligomers enhances catalytic activity and stability of clinical variants.
J.Biol.Chem., 298, 2022
|
|
8EUA
 
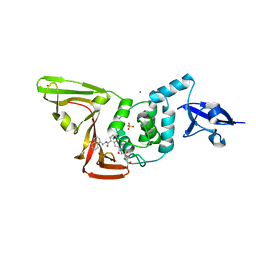 | | Structure of SARS-CoV2 PLpro bound to a covalent inhibitor | | Descriptor: | Papain-like protease nsp3, SULFATE ION, ZINC ION, ... | | Authors: | Mathews, I.I, Pokhrel, S, Wakatsuki, S. | | Deposit date: | 2022-10-18 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2.
Nat Commun, 14, 2023
|
|
6NA4
 
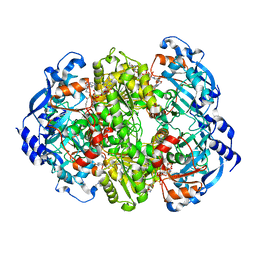 | | Co crystal structure of ECR with Butryl-CoA | | Descriptor: | 9-ETHYL-9H-PURIN-6-YLAMINE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | DeMirci, H. | | Deposit date: | 2018-12-05 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Intersubunit Coupling Enables Fast CO2-Fixation by Reductive Carboxylases
Acs Cent.Sci., 2022
|
|
6NA3
 
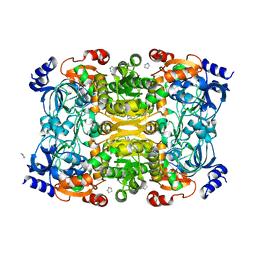 | | Crystal Structure of Apo-form of ECR | | Descriptor: | CHLORIDE ION, Putative crotonyl-CoA reductase, Pyrrolidine | | Authors: | DeMirci, H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intersubunit Coupling Enables Fast CO2-Fixation by Reductive Carboxylases
Acs Cent.Sci., 2022
|
|
6NA6
 
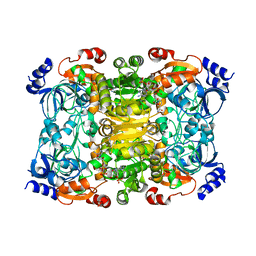 | |
6NA5
 
 | | Crystal Structure of ECR in complex with NADP+ | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative crotonyl-CoA reductase | | Authors: | DeMirci, H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intersubunit Coupling Enables Fast CO2-Fixation by Reductive Carboxylases
Acs Cent.Sci., 2022
|
|
7LS5
 
 | | Cryo-EM structure of the Pre3-1 20S proteasome core particle | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Schnell, H.M, Walsh Jr, R.M, Rawson, S, Hanna, J.W. | | Deposit date: | 2021-02-17 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structures of chaperone-associated assembly intermediates reveal coordinated mechanisms of proteasome biogenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LS6
 
 | | Cryo-EM structure of Pre-15S proteasome core particle assembly intermediate purified from Pre3-1 proteasome mutant (G34D) | | Descriptor: | Proteasome assembly chaperone 2, Proteasome chaperone 1, Proteasome maturation factor UMP1, ... | | Authors: | Schnell, H.M, Walsh Jr, R.M, Rawson, S, Hanna, J.W. | | Deposit date: | 2021-02-17 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structures of chaperone-associated assembly intermediates reveal coordinated mechanisms of proteasome biogenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LSX
 
 | | Cryo-EM structure of 13S proteasome core particle assembly intermediate purified from Pre3-1 proteasome mutant (G34D) | | Descriptor: | Proteasome assembly chaperone 2, Proteasome chaperone 1, Proteasome maturation factor UMP1, ... | | Authors: | Schnell, H.M, Walsh Jr, R.M, Rawson, S, Hanna, J.W. | | Deposit date: | 2021-02-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structures of chaperone-associated assembly intermediates reveal coordinated mechanisms of proteasome biogenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1SY8
 
 | | Structure of DNA sequence d-TGATCA by two-dimensional nuclear magnetic resonance spec and restrained molecular dynamics | | Descriptor: | 5'-D(P*TP*GP*AP*TP*CP*A)-3' | | Authors: | Barthwal, R, Awasthi, P, Narang, M, Sharma, U, Srivastava, N. | | Deposit date: | 2004-04-01 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of DNA sequence d-TGATCA by two-dimensional nuclear magnetic resonance spectroscopy and restrained molecular dynamics
J.STRUCT.BIOL., 148, 2004
|
|
