2E3Q
 
 | | Crystal structure of CERT START domain in complex with C18-ceramide (P212121) | | Descriptor: | DIMETHYL SULFOXIDE, Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2EAD
 
 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-fucosidase, CALCIUM ION, ... | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
2E3P
 
 | | Crystal structure of CERT START domain in complex with C16-cearmide (P1) | | Descriptor: | Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E3M
 
 | | Crystal structure of CERT START domain | | Descriptor: | Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2DX5
 
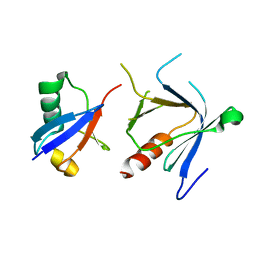 | | The complex structure between the mouse EAP45-GLUE domain and ubiquitin | | Descriptor: | Ubiquitin, Vacuolar protein sorting protein 36 | | Authors: | Hirano, S, Suzuki, N, Slagsvold, T, Kawasaki, M, Trambaiolo, D, Kato, R, Stenmark, H, Wakatsuki, S. | | Deposit date: | 2006-08-24 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain
Nat.Struct.Mol.Biol., 13, 2006
|
|
2DY3
 
 | | Crystal Structure of alanine racemase from Corynebacterium glutamicum | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Miyaguchi, I, Sasaki, C, Kato, R, Oikawa, T, Sugio, S. | | Deposit date: | 2006-09-05 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Alanine Racemase from Corynebacterium glutamicum at 2.1 A resolution
To be Published
|
|
2EAE
 
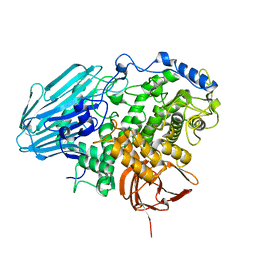 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complexes with products | | Descriptor: | Alpha-fucosidase, CALCIUM ION, alpha-L-fucopyranose, ... | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
2EAB
 
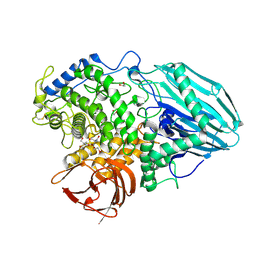 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-fucosidase, ... | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
2E3N
 
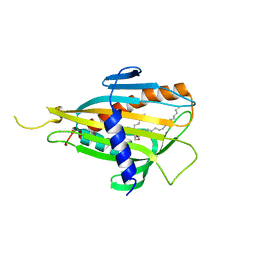 | | Crystal structure of CERT START domain in complex with C6-ceramide (P212121) | | Descriptor: | Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)HEXANAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E3R
 
 | | Crystal structure of CERT START domain in complex with C18-ceramide (P1) | | Descriptor: | Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E6V
 
 | | Crystal structure of VIP36 exoplasmic/lumenal domain, Ca2+/Man3GlcNAc-bound form | | Descriptor: | CALCIUM ION, Vesicular integral-membrane protein VIP36, alpha-D-mannopyranose, ... | | Authors: | Satoh, T, Kato, R, Wakatsuki, S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
2EAC
 
 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, Alpha-fucosidase, CALCIUM ION | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
2EAL
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with Forssman pentasaccharide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, Galectin-9 | | Authors: | Nagae, M, Nakamura-Tsuruta, S, Nishi, N, Nakamura, T, Hirabayashi, J, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-09-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the human galectin-9 N-terminal carbohydrate recognition domain reveals unexpected properties that differ from the mouse orthologue.
J.Mol.Biol., 375, 2008
|
|
2E3S
 
 | | Crystal structure of CERT START domain co-crystallized with C24-ceramide (P21) | | Descriptor: | Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2EAK
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with lactose | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, Galectin-9, ... | | Authors: | Nagae, M, Nakamura-Tsuruta, S, Nishi, N, Nakamura, T, Hirabayashi, J, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural analysis of the human galectin-9 N-terminal carbohydrate recognition domain reveals unexpected properties that differ from the mouse orthologue.
J.Mol.Biol., 375, 2008
|
|
2E3O
 
 | | Crystal structure of CERT START domain in complex with C16-ceramide (P212121) | | Descriptor: | DIMETHYL SULFOXIDE, Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3VTW
 
 | | Crystal structure of T7-tagged Optineurin LIR-fused human LC3B_2-119 | | Descriptor: | Optineurin, microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3VV1
 
 | | Crystal Structure of Caenorhabditis elegans galectin LEC-6 | | Descriptor: | MAGNESIUM ION, Protein LEC-6, beta-D-galactopyranose-(1-4)-alpha-L-fucopyranose | | Authors: | Makyio, H, Takeuchi, T, Tamura, M, Nishiyama, K, Takahashi, H, Natsugari, H, Arata, Y, Kasai, K, Yamada, Y, Wakatsuki, S, Kato, R. | | Deposit date: | 2012-07-10 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of preferential binding of fucose-containing saccharide by the Caenorhabditis elegans galectin LEC-6
Glycobiology, 23, 2013
|
|
3VIA
 
 | |
3VTU
 
 | | Crystal structure of human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3VHX
 
 | | The crystal structure of Arf6-MKLP1 (Mitotic kinesin-like protein 1) complex | | Descriptor: | ADP-ribosylation factor 6, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Makyio, H, Takei, T, Ohgi, H, Takahashi, S, Takatsu, H, Ueda, T, Kanaho, Y, Xie, Y, Shin, H.W, Kamikubo, H, Kataoka, M, Kawasaki, M, Kato, R, Wakatsuki, S, Nakayama, K. | | Deposit date: | 2011-09-12 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for Arf6-MKLP1 complex formation on the Flemming body responsible for cytokinesis
Embo J., 31, 2012
|
|
3VVH
 
 | |
3VTV
 
 | | Crystal structure of Optineurin LIR-fused human LC3B_2-119 | | Descriptor: | Optineurin, microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Suzuki, H, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-06-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for phosphorylation-triggered autophagic clearance of Salmonella
Biochem.J., 454, 2013
|
|
3WAL
 
 | | Crystal structure of human LC3A_2-121 | | Descriptor: | D-MALATE, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Suzuki, H, Tabata, K, Morita, E, Kawasaki, M, Kato, R, Dobson, R.C.J, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism.
Structure, 22, 2014
|
|
3WG1
 
 | | Crystal structure of Agrocybe cylindracea galectin with lactose | | Descriptor: | Galactoside-binding lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kuwabara, N, Hu, D, Tateno, H, Makio, H, Hirabayashi, J, Kato, R. | | Deposit date: | 2013-07-25 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational change of a unique sequence in a fungal galectin from Agrocybe cylindracea controls glycan ligand-binding specificity.
Febs Lett., 587, 2013
|
|
