2EDC
 
 | |
2EDA
 
 | |
2DIJ
 
 | | COMPLEX OF A Y195F MUTANT CGTASE FROM B. CIRCULANS STRAIN 251 COMPLEXED WITH A MALTONONAOSE INHIBITOR AT PH 9.8 OBTAINED AFTER SOAKING THE CRYSTAL WITH ACARBOSE AND MALTOHEXAOSE | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Strokopytov, B.V, Knegtel, R.M.A, Uitdehaag, J.C.M, Dijkstra, B.W. | | Deposit date: | 1998-05-27 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of cyclodextrin glycosyltransferase complexed with a maltononaose inhibitor at 2.6 angstrom resolution. Implications for product specificity.
Biochemistry, 35, 1996
|
|
2ORA
 
 | | RHODANESE (THIOSULFATE: CYANIDE SULFURTRANSFERASE) | | Descriptor: | OXIDIZED RHODANESE | | Authors: | Gliubich, F, Gazerro, M, Zanotti, G, Delbono, S, Berni, R. | | Deposit date: | 1996-02-22 | | Release date: | 1996-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Active site structural features for chemically modified forms of rhodanese.
J.Biol.Chem., 271, 1996
|
|
2PHH
 
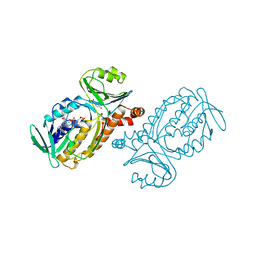 | |
2CXG
 
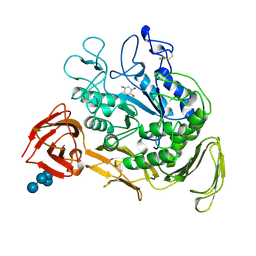 | | CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED TO THE INHIBITOR ACARBOSE | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Strokopytov, B.V, Uitdehaag, J.C.M, Ruiterkamp, R, Dijkstra, B.W. | | Deposit date: | 1998-05-08 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of cyclodextrin glycosyltransferase complexed with acarbose. Implications for the catalytic mechanism of glycosidases.
Biochemistry, 34, 1995
|
|
1TSI
 
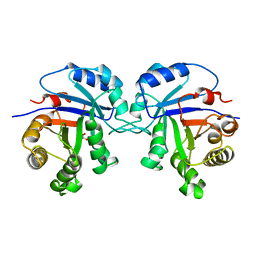 | |
4PAD
 
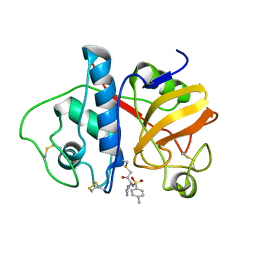 | |
1EAE
 
 | |
1EAD
 
 | |
1EAB
 
 | |
1EAA
 
 | |
1EAC
 
 | |
1EAF
 
 | |
1PBD
 
 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Mattevi, A, Hol, W.G.J. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBF
 
 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBB
 
 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2,4-DIHYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Mattevi, A, Hol, W.G.J. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBC
 
 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Van Der Bolt, F.J.T, Van Berkel, W.J.H. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1TCM
 
 | |
2X0N
 
 | | Structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei determined from Laue data | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, GLYCOSOMAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Vellieux, F.M.D, Hajdu, J, Hol, W.G.J. | | Deposit date: | 2009-12-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma Brucei Determined from Laue Data.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1SKZ
 
 | | PROTEASE INHIBITOR | | Descriptor: | ANTISTASIN, CHLORIDE ION | | Authors: | Krengel, U, Dijkstra, B.W. | | Deposit date: | 1997-04-16 | | Release date: | 1997-10-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of antistasin at 1.9 A resolution and its modelled complex with blood coagulation factor Xa.
EMBO J., 16, 1997
|
|
1B6G
 
 | | HALOALKANE DEHALOGENASE AT PH 5.0 CONTAINING CHLORIDE | | Descriptor: | CHLORIDE ION, GLYCEROL, HALOALKANE DEHALOGENASE, ... | | Authors: | Ridder, I.S, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-01-14 | | Release date: | 1999-07-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Haloalkane dehalogenase from Xanthobacter autotrophicus GJ10 refined at 1.15 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BGN
 
 | | P-HYDROXYBENZOATE HYDROXYLASE (PHBH) MUTANT WITH CYS 116 REPLACED BY SER (C116S) AND ARG 269 REPLACED BY THR (R269T), IN COMPLEX WITH FAD AND 4-HYDROXYBENZOIC ACID | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Eppink, M.H.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1998-05-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interdomain binding of NADPH in p-hydroxybenzoate hydroxylase as suggested by kinetic, crystallographic and modeling studies of histidine 162 and arginine 269 variants.
J.Biol.Chem., 273, 1998
|
|
1BGJ
 
 | | P-HYDROXYBENZOATE HYDROXYLASE (PHBH) MUTANT WITH CYS 116 REPLACED BY SER (C116S) AND HIS 162 REPLACED BY ARG (H162R), IN COMPLEX WITH FAD AND 4-HYDROXYBENZOIC ACID | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Eppink, M.H.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1998-05-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interdomain binding of NADPH in p-hydroxybenzoate hydroxylase as suggested by kinetic, crystallographic and modeling studies of histidine 162 and arginine 269 variants.
J.Biol.Chem., 273, 1998
|
|
1BF3
 
 | | P-HYDROXYBENZOATE HYDROXYLASE (PHBH) MUTANT WITH CYS 116 REPLACED BY SER (C116S) AND ARG 42 REPLACED BY LYS (R42K), IN COMPLEX WITH FAD AND 4-HYDROXYBENZOIC ACID | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Eppink, M.H.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1998-05-26 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lys42 and Ser42 variants of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens reveal that Arg42 is essential for NADPH binding.
Eur.J.Biochem., 253, 1998
|
|
