5SGI
 
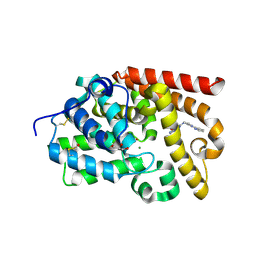 | | Crystal Structure of human phosphodiesterase 10 in complex with 6-(3-methylphenoxy)-2-(4-pyridin-2-ylpiperazin-1-yl)-9H-purine | | Descriptor: | 6-(3-methylphenoxy)-2-[4-(pyridin-2-yl)piperazin-1-yl]-9H-purine, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Brunner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
1NCT
 
 | | TITIN MODULE M5, N-TERMINALLY EXTENDED, NMR | | Descriptor: | TITIN | | Authors: | Pfuhl, M, Pastore, A. | | Deposit date: | 1996-08-13 | | Release date: | 1996-11-08 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | When a module is also a domain: the role of the N terminus in the stability and the dynamics of immunoglobulin domains from titin.
J.Mol.Biol., 265, 1997
|
|
1NCU
 
 | | Titin Module M5, N-terminally Extended, NMR | | Descriptor: | TITIN | | Authors: | Pfuhl, M, Pastore, A. | | Deposit date: | 1996-08-13 | | Release date: | 1996-11-08 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | When a module is also a domain: the role of the N terminus in the stability and the dynamics of immunoglobulin domains from titin.
J.Mol.Biol., 265, 1997
|
|
5EZI
 
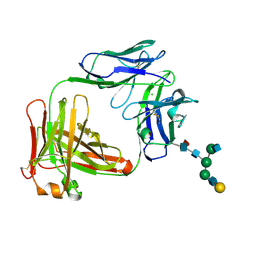 | | Crystal Structure of Fab of parasite invasion inhibitory antibody c1 - hexagonal form | | Descriptor: | CHLORIDE ION, Fab c12 Light chain, Fab c12 heavy chain, ... | | Authors: | Favuzza, P, Pluschke, G, Rudolph, M.G. | | Deposit date: | 2015-11-26 | | Release date: | 2017-01-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of the malaria vaccine candidate antigen CyRPA and its complex with a parasite invasion inhibitory antibody.
Elife, 6, 2017
|
|
5EZJ
 
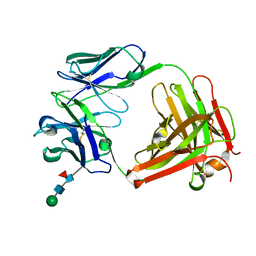 | |
5EZO
 
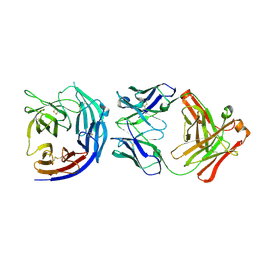 | | Crystal Structure of PfCyRPA in complex with an invasion-inhibitory antibody Fab. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PfCyRPA, c12 FAB, ... | | Authors: | Favuzza, P, Pluschke, G, Rudolph, M.G. | | Deposit date: | 2015-11-26 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Structure of the malaria vaccine candidate antigen CyRPA and its complex with a parasite invasion inhibitory antibody.
Elife, 6, 2017
|
|
5EZL
 
 | |
5EZN
 
 | | Crystal Structure of PfCyRPA | | Descriptor: | Cysteine-rich protective antigen | | Authors: | Favuzza, P, Pluschke, G, Rudolph, M.G. | | Deposit date: | 2015-11-26 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of the malaria vaccine candidate antigen CyRPA and its complex with a parasite invasion inhibitory antibody.
Elife, 6, 2017
|
|
5F03
 
 | | TRYPTASE B2 IN COMPLEX WITH 5-(3-Aminomethyl-phenoxymethyl)-3-[3-(2-chloro-pyridin-3-ylethynyl)-phenyl]-oxazolidin-2-one; compound with trifluoro-acetic acid | | Descriptor: | (5~{S})-5-[[3-(aminomethyl)phenoxy]methyl]-3-[3-[2-(2-chloranylpyridin-3-yl)ethynyl]phenyl]-1,3-oxazolidin-2-one, Tryptase beta-2 | | Authors: | Banner, D, Benz, J, Joseph, C, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
4ZGX
 
 | | Structure of aldosterone synthase (CYP11B2) in complex with (+)-(R)-N-(4-(4-chloro-3-fluorophenyl)-5,6,7,8-tetrahydroisoquinolin-8-yl)propionamide | | Descriptor: | Cytochrome P450 11B2, mitochondrial, N-[(8R)-4-(4-chloro-3-fluorophenyl)-5,6,7,8-tetrahydroisoquinolin-8-yl]propanamide, ... | | Authors: | Kuglstatter, A, Joseph, C. | | Deposit date: | 2015-04-24 | | Release date: | 2015-10-07 | | Last modified: | 2015-11-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of 4-Aryl-5,6,7,8-tetrahydroisoquinolines as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors: In Vivo Evaluation in Rodents and Cynomolgus Monkeys.
J.Med.Chem., 58, 2015
|
|
6XZ9
 
 | | Structure of aldosterone synthase (CYP11B2) in complex with 5-chloro-3,3-dimethyl-2-[5-[1-(1-methylpyrazole-4-carbonyl)azetidin-3-yl]oxy-3-pyridyl]isoindolin-1-one | | Descriptor: | 5-chloranyl-3,3-dimethyl-2-[5-[1-(1-methylpyrazol-4-yl)carbonylazetidin-3-yl]oxypyridin-3-yl]isoindol-1-one, Cytochrome P450 11B2, mitochondrial, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
1TNM
 
 | |
1TNN
 
 | |
4EYW
 
 | | Crystal structure of rat carnitine palmitoyltransferase 2 in complex with 1-[(R)-2-(3,4-Dihydro-1H-isoquinoline-2-carbonyl)-piperidin-1-yl]-2-phenoxy-ethanone | | Descriptor: | 1-[(2R)-2-(3,4-dihydroisoquinolin-2(1H)-ylcarbonyl)piperidin-1-yl]-2-phenoxyethanone, Carnitine O-palmitoyltransferase 2, mitochondrial, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Rufer, A, Joseph, C. | | Deposit date: | 2012-05-02 | | Release date: | 2013-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | Isothermal titration calorimetry with micelles: Thermodynamics of inhibitor binding to carnitine palmitoyltransferase 2 membrane protein.
FEBS Open Bio, 3, 2013
|
|
6XZ8
 
 | | Structure of aldosterone synthase (CYP11B2) in complex with N-[(1R)-1-[5-(6-chloro-1,1-dimethyl-3-oxo-isoindolin-2-yl)-3-pyridyl]ethyl]methanesulfonamide | | Descriptor: | Cytochrome P450 11B2, mitochondrial, HEME C, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
5Q0I
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-[(3,4-dimethoxyphenyl)-(4-methylphenyl)sulfonyl-amino]-N-(2,4-dimethylpentan-3-yl)ethanamide, Bile acid receptor, COACTIVATOR PEPTIDE PGC-1A PPAR GAMMA COACTIVATOR | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0Q
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, ethyl 4-({2-phenyl-5-[(thiophen-2-yl)sulfonyl]-4,5,6,7-tetrahydro-2H-pyrazolo[4,3-c]pyridine-3-carbonyl}amino)benzoate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1B
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-{[(2S)-2-cyclohexyl-2-{5,6-difluoro-2-[4-(1,3-thiazol-2-yl)phenyl]-1H-benzimidazol-1-yl}acetyl]amino}benzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q13
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-[6-chloro-2-(4-chlorophenyl)-5-fluoro-1H-benzimidazol-1-yl]-N-cyclohexyl-2-[(2S)-oxan-2-yl]acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0M
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 5-{[(3beta,5beta,14beta,17alpha)-3-hydroxy-24-oxocholan-24-yl]amino}benzene-1,3-dicarboxylic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0W
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-({5-bromo-1'-[(2-chlorophenyl)sulfonyl]-2-oxospiro[indole-3,4'-piperidin]-1(2H)-yl}methyl)benzoic acid, Bile acid receptor, cDNA FLJ76652, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q12
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-(2,6-difluorophenyl)-N-(2,6-dimethylphenyl)-5-methylimidazo[1,2-a]pyridin-3-amine, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1G
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2E)-N-cyclohexyl-N-(cyclohexylcarbamoyl)-3-(4-fluorophenyl)prop-2-enamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0U
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, trans-4-({(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylacetyl}amino)cyclohexyl hydrogen sulfate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1C
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-cyclohexyl-2-[2-(2,6-dimethoxypyridin-3-yl)-5,6-difluoro-1H-benzimidazol-1-yl]-N-(trans-4-hydroxycyclohexyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
